Integration of spatial protein imaging and transcriptomics in the human kidney tracks the regenerative potential of proximal tubules
- PMID: 40815665
- PMCID: PMC12356270
- DOI: 10.1126/sciadv.adv8918
Integration of spatial protein imaging and transcriptomics in the human kidney tracks the regenerative potential of proximal tubules
Abstract
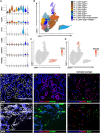
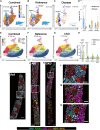
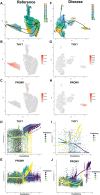
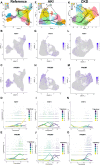
The organizational principles of nephronal segments are based on anatomical and physiological attributes that are linked to the homeostatic functions of the kidney. Recent molecular approaches have uncovered layers of deeper signatures and states in tubular cells that arise at various time points on the disease trajectory. Here, we introduce an analytical pipeline of multiplexed spatial protein imaging integrated with RNA expression to characterize proximal tubular subpopulations and neighborhoods in human kidney tissue. We demonstrate that, in reference tissue, a large proportion of S1 proximal tubular epithelial cells expresses thymus antigen 1 (THY1), a mesenchymal stromal and stem cell marker that regulates differentiation. Kidney disease is associated with loss of THY1 and transition toward expression of prominin 1 (PROM1), another stem cell marker recently linked to failed repair. Our data support a model in which the interplay between THY1 and PROM1 expression in proximal tubules associates with their regenerative potential and marks the timeline of disease progression.
Figures





Update of
-
Integration of spatial multiplexed protein imaging and transcriptomics in the human kidney tracks the regenerative potential timeline of proximal tubules.bioRxiv [Preprint]. 2024 Dec 2:2024.11.26.625544. doi: 10.1101/2024.11.26.625544. bioRxiv. 2024. Update in: Sci Adv. 2025 Aug 15;11(33):eadv8918. doi: 10.1126/sciadv.adv8918. PMID: 39677736 Free PMC article. Updated. Preprint.
References
-
- Lake B. B., Menon R., Winfree S., Hu Q., Melo Ferreira R., Kalhor K., Barwinska D., Otto E. A., Ferkowicz M., Diep D., Plongthongkum N., Knoten A., Urata S., Mariani L. H., Naik A. S., Eddy S., Zhang B., Wu Y., Salamon D., Williams J. C., Wang X., Balderrama K. S., Hoover P. J., Murray E., Marshall J. L., Noel T., Vijayan A., Hartman A., Chen F., Waikar S. S., Rosas S. E., Wilson F. P., Palevsky P. M., Kiryluk K., Sedor J. R., Toto R. D., Parikh C. R., Kim E. H., Satija R., Greka A., Macosko E. Z., Kharchenko P. V., Gaut J. P., Hodgin J. B., KPMP Consortium, Eadon M. T., Dagher P. C., El-Achkar T. M., Zhang K., Kretzler M., Jain S., An atlas of healthy and injured cell states and niches in the human kidney. Nature 619, 585–594 (2023). - PMC - PubMed
-
- Abedini A., Levinsohn J., Klotzer K. A., Dumoulin B., Ma Z., Frederick J., Dhillon P., Balzer M. S., Shrestha R., Liu H., Vitale S., Bergeson A. M., Devalaraja-Narashimha K., Grandi P., Bhattacharyya T., Hu E., Pullen S. S., Boustany-Kari C. M., Guarnieri P., Karihaloo A., Traum D., Yan H., Coleman K., Palmer M., Sarov-Blat L., Morton L., Hunter C. A., Kaestner K. H., Li M., Susztak K., Single-cell multi-omic and spatial profiling of human kidneys implicates the fibrotic microenvironment in kidney disease progression. Nat. Genet. 56, 1712–1724 (2024). - PMC - PubMed
-
- Canela V. H., Bowen W. S., Ferreira R. M., Syed F., Lingeman J. E., Sabo A. R., Barwinska D., Winfree S., Lake B. B., Cheng Y. H., Gaut J. P., Ferkowicz M., LaFavers K. A., Zhang K., Coe F. L., Worcester E., the Kidney Precision Medicine Project, Jain S., Eadon M. T., Williams J. C. Jr., El-Achkar T. M., A spatially anchored transcriptomic atlas of the human kidney papilla identifies significant immune injury in patients with stone disease. Nat. Commun. 14, 4140 (2023). - PMC - PubMed
MeSH terms
Substances
Grants and funding
- U01 DK114866/DK/NIDDK NIH HHS/United States
- U01 DK133097/DK/NIDDK NIH HHS/United States
- U01 DK114933/DK/NIDDK NIH HHS/United States
- U01 DK114908/DK/NIDDK NIH HHS/United States
- U01 DK133095/DK/NIDDK NIH HHS/United States
- U01 DK114907/DK/NIDDK NIH HHS/United States
- U54 DK137328/DK/NIDDK NIH HHS/United States
- U24 DK114886/DK/NIDDK NIH HHS/United States
- U01 DK114923/DK/NIDDK NIH HHS/United States
- U01 DK133113/DK/NIDDK NIH HHS/United States
- U01 DK133090/DK/NIDDK NIH HHS/United States
- UH3 DK114915/DK/NIDDK NIH HHS/United States
- U01 DK133768/DK/NIDDK NIH HHS/United States
- UH3 DK114861/DK/NIDDK NIH HHS/United States
- U01 DK133092/DK/NIDDK NIH HHS/United States
- UH3 DK114937/DK/NIDDK NIH HHS/United States
- U01 DK133081/DK/NIDDK NIH HHS/United States
- UH3 DK114926/DK/NIDDK NIH HHS/United States
- U01 DK133091/DK/NIDDK NIH HHS/United States
- U01 DK133093/DK/NIDDK NIH HHS/United States
- U54 DK134301/DK/NIDDK NIH HHS/United States
- U01 DK114920/DK/NIDDK NIH HHS/United States
- U01 DK133766/DK/NIDDK NIH HHS/United States
- OT2 OD033753/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

