Molecular epidemiology, antimicrobial resistance, and virulence characteristics of predominant methicillin-resistant Staphylococcus aureus clones with strong biofilm-producing capability from a tertiary teaching hospital in China
- PMID: 40817043
- PMCID: PMC12355878
- DOI: 10.1186/s12866-025-04258-z
Molecular epidemiology, antimicrobial resistance, and virulence characteristics of predominant methicillin-resistant Staphylococcus aureus clones with strong biofilm-producing capability from a tertiary teaching hospital in China
Abstract
Background: Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most prevalent bacterial pathogens leading to various kinds of infections, but the characteristics of this superbug with both strong biofilm-producing and intracellular invasive capabilities is rarely reported. This study aimed to investigate the genotypic and phenotypic features of this superbug with above two properties.
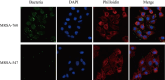
Methods: Phenotypic resistance profiling of MRSA clinical isolates was performed via the VITEK 2 AST-GP67 Test Kit. Biofilm production was assessed via crystal violet staining and the Congo red agar (CRA) method. The biofilm-degrading activity was tested using Proteinase K, Dispersin B, and DNase I. The intracellular invasive capability was evaluated via dilution plate count and immunofluorescence assay. Genotyping was performed using multilocus sequence typing and staphylococcal protein A typing methods, and virulence genes were detected via polymerase chain reaction. Flow cytometry was performed to assess the cytotoxicity of the dominant MRSA clones.
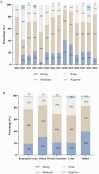
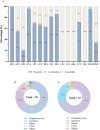
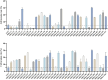
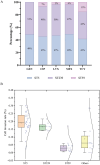
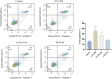
Results: A high prevalence (21.6%) of MRSA isolates exhibiting strong biofilm-forming capability was observed in this study, including 70 strains with the highest level of biofilm production (optical density > 0.4). DNase I exhibited the most effective biofilm-degrading activity, with the biofilm-degrading percentage of 78.6% of the strains exceeding 50%. Simultaneously, 71.4% of the isolates exhibited strong invasive capability into A549 cells. ST5-t2460 (48.6%), ST59-t437 (20%), and ST239-t030 (11.4%) were identified as the predominant clones. In particular, ST5-t2460 and ST239-t030 clones exhibited broader antibiotic resistance to gentamicin, ciprofloxacin, levofloxacin, moxifloxacin, and tetracycline compared with ST59-t437 clone. In addition, a higher percentage of the isolates belonging to ST5-t2460 (91.2%) and ST239-t030 (100%) clones demonstrated stronger intracellular invasive capability relative to those belonging to ST59-t437 clone (14.3%). Furthermore, ST5-t2460 and ST239-t030 clones displayed stronger cytotoxicity and carried higher proportions of adhesion-related genes (fnbA, sdrD, sasC) and other virulence genes (sea, seb, sec, isdB, lukE-D, tsst-1).
Conclusions: This is the first report of the phenotypic-genotypic characteristics of MRSA with both strong biofilm-producing and virulence potential, with ST5-t2460, ST59-t437, and ST239-t030 clones accounting for the major genotypes. Further exploration of specific virulence genes correlating to the pathogenesis of this superbug is deemed essential for developing targeted infection control and treatment strategies in the future.
Keywords: Biofilm; Epidemiology; Infection control; MRSA; Molecular characterization; Virulence gene.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study complies with the Declaration of Helsinki. The bacterial strains used in this study were isolated from the routine biological specimens, which were obtained during the clinical diagnosis and management of the patients. Also, rights and health of the subjects were not under threat, and no personal identifying information was used during this study. The mutated isolates were destroyed using autoclaving device and finally processed as medical wastes, according to the regulation of Affiliated hospital of Inner Mongolian Medical University. According to the national regulation on ethical review (No. 2016–11, 12/01/2016), the requirement for the informed consent was waived by the Ethical Committee of Inner Mongolian Medical University, which belongs to the Office of Scientific Research of Inner Mongolian Medical University. Meanwhile, this study was approved by the Ethical Committee of the Inner Mongolian Medical University (Reference No. YKD202201166). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Antimicrobial resistance and molecular characteristics of bovine mastitis-associated methicillin-resistant Staphylococcus aureus: potential for cross-species transmission of ST59-MRSA.Microbiol Spectr. 2025 Jul;13(7):e0280024. doi: 10.1128/spectrum.02800-24. Epub 2025 May 21. Microbiol Spectr. 2025. PMID: 40396723 Free PMC article.
-
A cross-sectional molecular epidemiological study of biofilm-producing methicillin-resistant Staphylococcus aureus.Medicine (Baltimore). 2025 Jul 18;104(29):e43346. doi: 10.1097/MD.0000000000043346. Medicine (Baltimore). 2025. PMID: 40696645 Free PMC article.
-
Phenotypic and genotypic characteristics of mecA - positive oxacillin-sensitive Staphylococcus aureus isolated from patients with bloodstream infection in a tertiary hospital in Southern Brazil.Braz J Microbiol. 2024 Sep;55(3):2705-2713. doi: 10.1007/s42770-024-01420-z. Epub 2024 Jun 19. Braz J Microbiol. 2024. PMID: 38896343 Free PMC article.
-
Nasal Staphylococcus aureus and S. pseudintermedius carriage in healthy dogs and cats: a systematic review of their antibiotic resistance, virulence and genetic lineages of zoonotic relevance.J Appl Microbiol. 2022 Dec;133(6):3368-3390. doi: 10.1111/jam.15803. Epub 2022 Sep 27. J Appl Microbiol. 2022. PMID: 36063061 Free PMC article.
-
Prevalence, trend and antimicrobial susceptibility of Methicillin Resistant Staphylococcus aureus in Nigeria: a systematic review.J Infect Public Health. 2018 Nov-Dec;11(6):763-770. doi: 10.1016/j.jiph.2018.05.013. Epub 2018 Jun 19. J Infect Public Health. 2018. PMID: 29933910
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

