Aging increases susceptibility to liver fibrosis through enhanced NAT10-mediated ac4C modification of TGFβ1 mRNA
- PMID: 40817062
- PMCID: PMC12355746
- DOI: 10.1186/s13073-025-01520-x
Aging increases susceptibility to liver fibrosis through enhanced NAT10-mediated ac4C modification of TGFβ1 mRNA
Abstract
Background: The epidemiological observational studies unveiled that aging is one of the risk factors for liver fibrosis, and the hepatic tissues in the elderly harbor more fibrotic lesions when compared to those in young people. Previous investigations found that TGFβ1 was elevated with aging and promoted liver fibrosis. However, the underlying mechanisms of aging and liver fibrosis remain largely unknown.
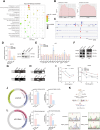
Methods: CCl4-induced liver fibrosis animal models were used in this study. The impact of NAT10 on liver fibrosis and cellular senescence was analyzed by using NAT10 overexpression or knockout hepatic stellate cell lines. The distribution of ac4C RNA modification was monitored by the acRIP-seq. The RNA-protein interaction was examined by the RNA immunoprecipitation.
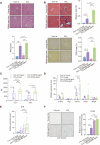
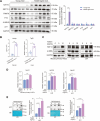
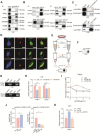
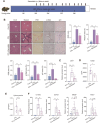
Results: We demonstrated that the middle-aged mice were more susceptible to the CCl4-induced liver fibrosis when compared to the young mice. Then, we found that RNA ac4C-modifying enzyme NAT10 was transcriptionally activated by TGFβ1/SMAD2/3 axis and highly expressed in the aging liver as well as liver fibrosis mouse model. Suppression of NAT10 by its inhibitor Remodelin or specific shRNA attenuated senescence and activation of hepatic stellate cells. Subsequent studies found that NAT10 directly triggered the ac4C RNA modification of TGFβ1 mRNA by physically interacting with the RNA-binding protein PTBP1, enhancing the stabilization of TGFβ1 mRNA and subsequent activation of TGFβ/SMAD signaling pathway. Animal studies demonstrated that inhibition of NAT10 by Remodelin significantly alleviated liver fibrosis and cellular senescence.
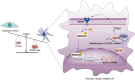
Conclusions: Our study identified a previously unknown mechanism of how TGFβ1 drives cellular senescence and liver fibrosis through NAT10-mediated ac4C mRNA modification.
Keywords: Aging; Liver fibrosis; NAT10; RNA modification; TGFβ/SMAD signaling pathway.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: Animals in this study were used in accordance with the Guide for Care and Use of Laboratory Animals of the National Institute of Health. The animal study was approved by the institutional review boards and ethics committees of The Third Affiliated Hospital of Sun Yat-sen University. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Sheedfar F, Di Biase S, Koonen D, Vinciguerra M. Liver diseases and aging: friends or foes? Aging Cell. 2013;12:950–4. - PubMed
-
- Caballeria L, Pera G, Arteaga I, Rodriguez L, Aluma A, Morillas RM, de la Ossa N, Diaz A, Exposito C, Miranda D, et al. High prevalence of liver fibrosis among European adults with unknown liver disease: a population-based study. Clin Gastroenterol Hepatol. 2018;16:1138-1145 e1135. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

