Population structure in a fungal human pathogen is potentially linked to pathogenicity
- PMID: 40817095
- PMCID: PMC12356954
- DOI: 10.1038/s41467-025-62777-9
Population structure in a fungal human pathogen is potentially linked to pathogenicity
Abstract
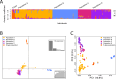
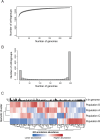
Aspergillus flavus is a clinically and agriculturally important saprotrophic fungus responsible for severe human infections and extensive crop losses. Here, we analyze genomic data from 300 (117 clinical and 183 environmental) A. flavus isolates from 13 countries, including 82 clinical isolates sequenced in this study, to examine population and pan-genome structure and their relationship to pathogenicity. We use single nucleotide polymorphisms to build a phylogeny, analyze admixture, and perform discriminant analysis of principal components. We identify five A. flavus populations, including a new population, D, corresponding to distinct clades in the genome-wide phylogeny. Strikingly, > 75% of clinical isolates were in population D and <5% in population B. We also use orthogroup clustering to identify core and accessory genes within the pan-genome. Accessory genes, including genes within biosynthetic gene clusters, were significantly more common in some populations but rare in others. Our functional annotations show that population D is enriched for genes associated with carbohydrate metabolism, lipid metabolism and certain types of hydrolase activity, whereas a non-clinical population is depleted in genes related to zinc ion binding. In contrast to previous results from the major human pathogen Aspergillus fumigatus, isolation of A. flavus from human specimens is associated with population structure, providing a promising system for future investigations into the contributions of population-specific genetic differences to human infection.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.R. is a scientific consultant for LifeMine Therapeutics, Inc. The other authors declare no other competing interests.
Figures


Update of
-
Pathogenicity is associated with population structure in a fungal pathogen of humans.bioRxiv [Preprint]. 2024 Jul 10:2024.07.05.602241. doi: 10.1101/2024.07.05.602241. bioRxiv. 2024. Update in: Nat Commun. 2025 Aug 15;16(1):7594. doi: 10.1038/s41467-025-62777-9. PMID: 39026826 Free PMC article. Updated. Preprint.
References
-
- Schauwvlieghe, A. F. A. D. et al. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respir. Med.6, 782–792 (2018). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

