Sepsis Induces Long-Term Muscle and Mitochondrial Dysfunction due to Autophagy Disruption Amenable by Urolithin A
- PMID: 40817441
- PMCID: PMC12356704
- DOI: 10.1002/jcsm.70041
Sepsis Induces Long-Term Muscle and Mitochondrial Dysfunction due to Autophagy Disruption Amenable by Urolithin A
Abstract
Background: Sepsis survivors often experience sustained muscle weakness, leading to physical disability, with no pharmacological treatments available. Despite these well-documented long-term clinical consequences, research exploring the cellular and molecular mechanisms is sorely lacking.
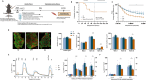
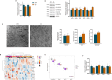
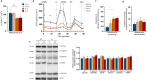
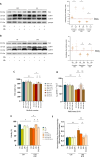
Methods: Bioinformatic analysis was performed in the vastus lateralis transcriptome of human ICU survivors 7 days after ICU discharge (D7), 6 months (M6) and age- and sex-matched controls. Enrichment analysis using Gene Ontology (GO) terms and Mitocarta3.0 was performed at D7 and M6 on differentially expressed genes (DEGs) and modules identified by weighted gene co-expression network analysis (WGCNA). Using a murine model of resuscitated sepsis induced by caecal slurry injection, pathways identified by the bioinformatics analysis were explored in 18- to 24-week-old sepsis-surviving (SS) mice at Day 10. Autophagy flux was investigated both in vivo and in vitro with chloroquine, a lysosomal inhibitor and urolithin A (UA), an autophagy inducer. Systemic metabolism was evaluated with indirect calorimetry, muscle phenotype with in situ and ex vivo contractility, muscle mass, myofibre cross-sectional area and typing and mitochondrial population with transmission electron microscopy (TEM), as well as mitochondrial function with high-resolution respirometry. Autophagic vacuole (AV) level was monitored using LC3B-II and P62 protein expression and TEM.
Results: Pathways related to 'mitochondrion' were the only ones whose deregulation persisted between D7 and M6 (p < 0.05) and characterized WGCNA modules correlated with muscle mass, strength and physical function. Shared mitochondrial DEGs between D7 and M6 encoded matrix mitochondrial proteins related to 'metabolism' and 'mitochondrial dynamics'. SS mice exhibited reduced complex I-driven oxygen consumption (CI-JO2) (-45%), increased S-nitrosylation of complex I, damaged (+35%) and oxidized (+51%) mitochondria and AV accumulation (5 vs. 50 AVs/mm2) compared with sham pair-fed mice (p < 0.05) despite no differences in mitochondrial size or number. Autophagy flux was reduced in SS mice due to decreased AV degradation ratio (p < 0.05). UA restored a balanced autophagy flux (turnover ratio 0.96 vs. -0.17) by increasing AVs formation and degradation ratio (p < 0.05). UA also improved CI-JO2 (81 vs. 106 pmol/s/mg), tetanic force (215 vs. 244 mN/mm2) and hindlimb muscle weight in SS mice (p < 0.05).
Conclusion: Mitochondrial and autophagy disruption contributes to long-term muscle dysfunction in human and mouse sepsis survivors. We demonstrate for the first time that sepsis induces an autophagy flux blockade. Urolithin A prevents mitochondrial and muscle impairments both in vivo and in vitro by improving autophagy flux.
Keywords: autophagy; mitochondria; muscle; sepsis.
© 2025 The Author(s). Journal of Cachexia, Sarcopenia and Muscle published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Poulsen J. B., Rose M. H., Jensen B. R., Møller K., and Perner A., “Biomechanical and Nonfunctional Assessment of Physical Capacity in Male ICU Survivors*,” Critical Care Medicine 41, no. 1 (2013): 93–101. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

