Maternal diet-induced alterations in uterine fluid sncRNAs compromise preimplantation embryo development and offspring metabolic health
- PMID: 40818970
- PMCID: PMC12357936
- DOI: 10.1038/s41467-025-63054-5
Maternal diet-induced alterations in uterine fluid sncRNAs compromise preimplantation embryo development and offspring metabolic health
Abstract
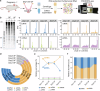
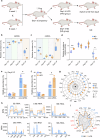
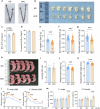
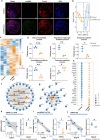
The periconception period is critical for embryo development, pregnancy outcomes, and offspring health. During this stage, oviductal and uterine fluids facilitate embryo-maternal interactions and support early embryonic development. Using PANDORA-seq, we identify a diverse repertoire of small non-coding RNAs in female mouse oviduct fluid and uterine fluid during preimplantation, with tRNA-derived small RNAs and rRNA-derived small RNAs being predominant. Maternal high-fat diet during preimplantation period significantly alters tsRNA and rsRNA expression in oviduct fluid and uterine fluid compared to normal diet, disrupting blastocyst metabolic gene expression. While implantation remained unaffected, these alterations impair mid-gestation embryonic and placental growth, resulting in reduced birth weight and length, as well as metabolic disorders in offspring. Furthermore, transfecting embryos with uterine fluid-derived sncRNAs altered by maternal high-fat diet mimics the in vivo effects. These findings suggest that tsRNAs and rsRNAs in reproductive fluids may reflect maternal metabolic status and transmit dietary information to the early embryo, which might influence pregnancy outcomes and offspring health.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Seminal plasma uterine priming alters uterine transcriptomics and negatively impacts embryo growth and uterine artery resistance but not offspring liver transcriptomics in beef cattle.J Anim Sci. 2024 Jan 3;102:skae300. doi: 10.1093/jas/skae300. J Anim Sci. 2024. PMID: 39367540
-
Supplementation of the maternal diet during pregnancy with chocolate and fructose interacts with the high-fat diet of the young to facilitate the onset of metabolic disorders in rat offspring.Clin Exp Pharmacol Physiol. 2013 Sep;40(9):652-61. doi: 10.1111/1440-1681.12147. Clin Exp Pharmacol Physiol. 2013. PMID: 23819696
-
One-carbon metabolites supplementation and nutrient restriction alter the fetal liver metabolomic profile during early gestation in beef heifers.J Anim Sci. 2024 Jan 3;102:skae258. doi: 10.1093/jas/skae258. J Anim Sci. 2024. PMID: 39234988
-
Clinical outcomes following selection of human preimplantation embryos with time-lapse monitoring: a systematic review.Hum Reprod Update. 2014 Sep-Oct;20(5):617-31. doi: 10.1093/humupd/dmu023. Epub 2014 Jun 2. Hum Reprod Update. 2014. PMID: 24890606
-
Association Between Human Embryo Culture Conditions, Cryopreservation, and the Potential Risk of Birth Defects in Children Conceived Through Assisted Reproduction Technology.Medicina (Kaunas). 2025 Jun 30;61(7):1194. doi: 10.3390/medicina61071194. Medicina (Kaunas). 2025. PMID: 40731824 Free PMC article. Review.
References
-
- Barker, D., Barker, M., Fleming, T. & Lampl, M. Developmental biology: support mothers to secure future public health. Nature504, 209–211 (2013). - PubMed
-
- Velazquez, M. A., Fleming, T. P. & Watkins, A. J. Periconceptional environment and the developmental origins of disease. J. Endocrinol.242, T33–T49 (2019). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

