Integrative machine learning models predict prostate cancer diagnosis and biochemical recurrence risk: Advancing precision oncology
- PMID: 40819002
- PMCID: PMC12357910
- DOI: 10.1038/s41746-025-01930-6
Integrative machine learning models predict prostate cancer diagnosis and biochemical recurrence risk: Advancing precision oncology
Abstract
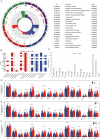
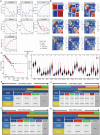
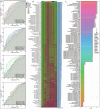
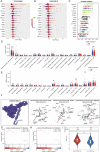
Prostate cancer (PCa) ranks among the most prevalent cancers in men worldwide. Biochemical recurrence (BCR) presents a major clinical challenge in PCa management, with significant prognostic heterogeneity observed among patients post-recurrence. This study aimed to develop machine learning models for predicting both the diagnosis and prognosis of PCa patients. Using WGCNA, we initially identified 16 BCR-related target genes. Cluster analysis revealed these genes were significantly associated with PCa prognosis, drug sensitivity, and immune infiltration. We constructed a robust diagnostic model integrating multiple machine learning algorithms, demonstrating strong predictive capability for PCa. Furthermore, a BCR-related prognostic model built using the LASSO algorithm also yielded satisfactory performance. Among the differentially expressed BCR-associated prognostic genes, COMP emerged as a critical regulatory factor. Both in vitro and in vivo experiments confirmed COMP's role in influencing PCa progression. Additionally, COMP demonstrates significant potential as a dual biomarker for both the diagnosis and recurrence prediction of PCa.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Cornford, P. et al. EAU-EANM-ESTRO-ESUR-ISUP-SIOG guidelines on prostate cancer-2024 update. Part I: screening, diagnosis, and local treatment with curative intent. Eur. Urol.86, 148–163 (2024). - PubMed
-
- Van den Broeck, T. et al. Prognostic value of biochemical recurrence following treatment with curative intent for prostate cancer: a systematic review. Eur. Urol.75, 967–987 (2019). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

