Human neural organoid microphysiological systems show the building blocks necessary for basic learning and memory
- PMID: 40819006
- PMCID: PMC12357958
- DOI: 10.1038/s42003-025-08632-5
Human neural organoid microphysiological systems show the building blocks necessary for basic learning and memory
Abstract
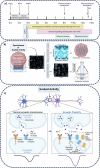
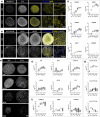
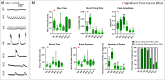
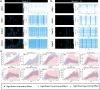
Brain Microphysiological Systems, including neural organoids derived from human induced pluripotent stem cells, offer a unique lens to study the intricate workings of the human brain. This paper investigates the foundational elements of learning and memory in neural organoids by quantifying immediate early gene expression in response to chemical modulation, input-specific short- and long-term synaptic plasticity, neuronal network dynamics, connectivity, and criticality to demonstrate the utility of these organoids in basic science research. Neural organoids showed synapse formation, glutamatergic and GABAergic receptor expression, immediate early gene expression basally and evoked, functional connectivity, criticality, and synaptic plasticity in response to theta-burst stimulation. In addition, pharmacological interventions on GABAergic and glutamatergic receptors and input-specific theta-burst stimulation further shed light on the capacity of neural organoids to mirror synaptic modulation, specifically short- and long-term potentiation and depression, demonstrating their potential as tools for studying neurophysiological and neurological processes and informing therapeutic strategies for diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: T.H. is named inventor on a patent by Johns Hopkins University on the production of organoids, which is licensed to Axo-Sim, New Orleans, LA, USA. T.H. and L.S. are consultants for AxoSim, New Orleans, and T.H. is also a consultant for AstraZeneca and American Type Culture Collection (ATCC) on advanced cell culture methods. B.J.K. is a named inventor on patents by CCLabs Pty Ltd trading as Cortical Labs on the use of biological neural systems for intelligent purposes. B.J.K., F.H, and A.L are employees of Cortical Labs. B.J.K. and A.L. are shareholders of Cortical Labs. J.L is a data science consultant for Vindhya Data Science specializing in bioinformatics analysis. The rest of the authors declare no conflict of interest.
Figures


Update of
-
Human Neural Organoid Microphysiological Systems Show the Building Blocks Necessary for Basic Learning and Memory.bioRxiv [Preprint]. 2024 Sep 19:2024.09.17.613333. doi: 10.1101/2024.09.17.613333. bioRxiv. 2024. Update in: Commun Biol. 2025 Aug 16;8(1):1237. doi: 10.1038/s42003-025-08632-5. PMID: 39345518 Free PMC article. Updated. Preprint.
References
-
- Smirnova, L. & Hartung, T. The promise and potential of brain organoids. Adv. Healthc. Mater. e2302745 10.1002/adhm.202302745 (2024). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

