Intron turnover of slc26a1 and slc26a2 and convergence of intron insertion sites
- PMID: 40819135
- PMCID: PMC12357937
- DOI: 10.1038/s41598-025-15147-w
Intron turnover of slc26a1 and slc26a2 and convergence of intron insertion sites
Abstract
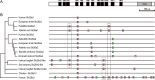
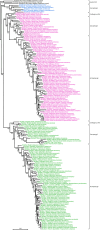
Intron gain and loss are rare events in vertebrates; however, comparative genome analysis of elephant sharks, tetrapods, and teleosts revealed a higher level of intron turnover in teleosts. slc26a1 and slc26a2 are members of the anion-exchanger gene family. Human, zebrafish, and Japanese pufferfish slc26a1 consist of two, two, and seven exons, respectively, and slc26a2, two, three, and four exons, respectively. To better understand intron turnover in teleosts, we analyzed the exon-intron organization of slc26a1 and slc26a2 in 81 vertebrates, including 62 ray-finned fish. In most Eurypterygii, which comprise the majority of the Neoteleostei and include Acanthomorpha, Aulopiformes, and Myctophiformes, slc26a1 and slc26a2 have seven and four exons, respectively, whereas those of most other ray-finned fishes consist of two and three exons, respectively, suggesting that intron gain occurred in both slc26a1 and slc26a2 of the Eurypterygii ancestor. In addition, notothenioid slc26a2 has six exons, suggesting that two introns were inserted into the notothenioid ancestor. The two newly acquired introns in the notothenioid consist of transposon-like sequences, suggesting that they were generated via transposon insertion. The positions of some of the newly acquired introns of slc26a1 and slc26a2 in Eurypterygii are identical or very close to those of other slc26 members. These results demonstrate the lineage-specific intron gains of slc26a1 and slc26a2 in ray-finned fish and convergence at the insertion sites of some of the newly acquired introns.
Keywords: Convergent evolution; Intron gain; Intron turnover; Ray-finned fish; Transposable element.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Topical versus systemic antibiotics for chronic suppurative otitis media.Cochrane Database Syst Rev. 2025 Jun 9;6(6):CD013053. doi: 10.1002/14651858.CD013053.pub3. Cochrane Database Syst Rev. 2025. PMID: 40484402
-
The quantity, quality and findings of network meta-analyses evaluating the effectiveness of GLP-1 RAs for weight loss: a scoping review.Health Technol Assess. 2025 Jun 25:1-73. doi: 10.3310/SKHT8119. Online ahead of print. Health Technol Assess. 2025. PMID: 40580049 Free PMC article.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
References
-
- Roy, S. W. & Gilbert, W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet.7, 211–221. 10.1038/nrg1807 (2006). - PubMed
-
- Matlin, A. J., Clark, F. & Smith, C. W. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol.6, 386–398. 10.1038/nrm1645 (2005). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

