Network pharmacology and metabolomics reveal mathurameha, a Thai traditional Anti-Diabetic formula, enhances glucose metabolism through PI3K-AKT/AMPK/GLUT4 pathway modulation
- PMID: 40820159
- PMCID: PMC12358555
- DOI: 10.1038/s41598-025-15556-x
Network pharmacology and metabolomics reveal mathurameha, a Thai traditional Anti-Diabetic formula, enhances glucose metabolism through PI3K-AKT/AMPK/GLUT4 pathway modulation
Abstract
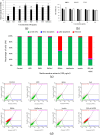
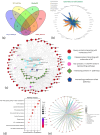
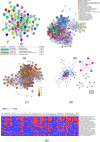
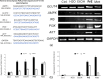
Traditional herbal formulations offer promising avenues for diabetes management by targeting multiple molecular pathways. Mathurameha (MT), a polyherbal preparation, has been historically used for its antidiabetic potential. However, its molecular mechanisms remain largely unexplored. FrE exhibited potent α-glucosidase inhibition (IC₅₀ 0.3 µg/mL) and significantly enhanced glucose uptake in L6 myotubes (3.67 ± 0.23-fold) and 3T3-L1 adipocytes (IC₅₀ 6.78 µg/mL). It also stimulated insulin secretion (1.42-fold), comparable to metformin (1.46-fold), and protected INS-1 pancreatic β-cells from H₂O₂-induced apoptosis (30.65 ± 3.54%) through partial caspase-3 inhibition. LC-MS-QTOF analysis identified 73 metabolites, including ellagic acid, kushenol A, gallic acid, arctiin, neoandrographolide, astilbin, paenol, muricatacin, coumarrayin, and zingerone. Network pharmacology and pathway enrichment analyses revealed key targets (GSK3β, GLUT4, PPARG, INSR, AKT2, CASP3, and MMP9) and highlighted the involvement of PI3K-AKT, AMPK, and GLUT4 signaling pathways. Gene expression analysis confirmed the upregulation of GLUT4, AMPK, IRS, PI3K, and AKT genes in L6 myotubes treated with FrE. These findings suggest that MT exerts antidiabetic effects via the PI3K-AKT/AMPK/GLUT4 signaling axis, promoting glucose uptake, insulin secretion, and β-cell protection. Future studies will focus on in vivo validation, standardization of bioactive fractions, and omics-based approaches to establish a well-defined, effective formulation for diabetes management.
Keywords: Antidiabetic activity; Glucose transport; Insulin secretion enrichment; LC-MS/MS-QTOF analysis; Mathurameha; Network pharmacology; Thai traditional medicine; Twenty-six medicinal plant mixture.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
A new chromanone isolated from Portulaca oleracea L. increases glucose uptake by stimulating GLUT4 translocation to the plasma membrane in 3T3-L1 adipocytes.Int J Biol Macromol. 2019 Feb 15;123:26-34. doi: 10.1016/j.ijbiomac.2018.10.206. Epub 2018 Oct 30. Int J Biol Macromol. 2019. PMID: 30389528
-
Chiisanogenin enhances glucose uptake and lowers blood glucose via insulin signaling activation.Biomed Pharmacother. 2025 Aug;189:118281. doi: 10.1016/j.biopha.2025.118281. Epub 2025 Jun 18. Biomed Pharmacother. 2025. PMID: 40554287
-
Integrated metabolomics and network pharmacology reveal the PI3K/Akt-mediated therapeutic mechanism of Abrus cantoniensis in lipid metabolism disorders.Phytomedicine. 2025 Sep;145:156953. doi: 10.1016/j.phymed.2025.156953. Epub 2025 Jun 9. Phytomedicine. 2025. PMID: 40517619
-
Antidiabetic Potential of Sophora Species: Mechanisms, Bioactive Constituents, and Therapeutic Prospects.Planta Med. 2025 Sep;91(10-11):546-557. doi: 10.1055/a-2597-8133. Epub 2025 Apr 30. Planta Med. 2025. PMID: 40306687 Review.
-
GLUT4 Trafficking and Storage Vesicles: Molecular Architecture, Regulatory Networks, and Their Disruption in Insulin Resistance.Int J Mol Sci. 2025 Aug 5;26(15):7568. doi: 10.3390/ijms26157568. Int J Mol Sci. 2025. PMID: 40806697 Free PMC article. Review.
References
-
- Dahlén, A. D. et al. Trends in antidiabetic drug discovery: FDA approved drugs, new drugs in clinical trials and global sales. Front. Pharmacol.1210.3389/fphar.2021.807548 (2022). https://www.frontiersin.org/articles/ (accessed 26 Nov2022). - PMC - PubMed
-
- Clapham, J. C. Sixty years of drug discovery for type 2 diabetes: where are we now?? In Type 2 Diabetes: Methods and Protocols (ed. Stocker, C. J.) 1–30 (Springer, 2020). - PubMed
MeSH terms
Substances
Grants and funding
- POSTDOCTORAL FELLOWSHIP , GRANT NO. 04/2025/Mae Fah Luang University
- DBG6280007/Thailand Science Research and Innovation Fund
- F01- 683R-17-046/Office of the Permanent Secretary, Ministry of Higher Education, Science, Research and Innovation, Thailand
- N35E680089/The Hub of Knowledge: Herbs for Sustainable Health and Well-being Consortium and supported by the National Research Council of Thailand
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

