Developing a Gram-Negative Selective Peptide-Drug Conjugate
- PMID: 40821530
- PMCID: PMC12355328
- DOI: 10.1021/acsomega.4c08000
Developing a Gram-Negative Selective Peptide-Drug Conjugate
Abstract
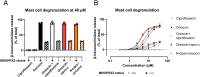
Resistance to fluoroquinolone antibiotics has serious implications for healthcare; here, we conjugate the widely used fluoroquinolone ciprofloxacin to a proline-rich antimicrobial peptide (PrAMP) oncocin to improve oncocin's potency in ciprofloxacin-sensitive and ciprofloxacin-resistant strains of Escherichia coli. The conjugate molecule (oncocin-cipro-c) is ∼3× more potent than the parent oncocin, as determined by MIC, while retaining Gram-negative selectivity. We have characterized oncocin-cipro-c's interactions with three intracellular targets, two from oncocin (DnaK and 70S ribosome) and a third from ciprofloxacin (gyrase). Oncocin-cipro-c is also able to facilitate mast cell degranulation at a lower concentration than the parent peptide. The development of multimode antibiotics like oncocin-cipro-c is essential in the coming decades of antibiotic resistance.
© 2025 The Authors. Published by American Chemical Society.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
