Deciphering the proteome of Escherichia coli K-12: Integrating transcriptomics and machine learning to annotate hypothetical proteins
- PMID: 40821719
- PMCID: PMC12356324
- DOI: 10.1016/j.csbj.2025.07.036
Deciphering the proteome of Escherichia coli K-12: Integrating transcriptomics and machine learning to annotate hypothetical proteins
Abstract
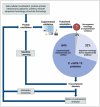
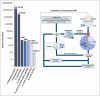
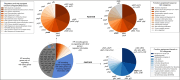
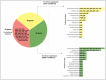
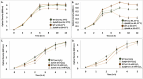
Omics technologies have led to the discovery of a vast number of proteins that are expressed but have no functional annotation - so called hypothetical proteins (HPs). Even in the best-studied model organism Escherichia coli K-12, over 2 % of the proteome remains uncharacterized. This knowledge gap becomes even worse when looking at microbial dark matter. However, knowing the functions of proteins is crucial for elucidating cellular and metabolic processes and harnessing biotechnological potentials. Here, we employed machine learning to decipher the transcriptional regulatory network of E. coli K-12, as well as other in silico tools to assign functions to uncharacterized HPs. We further provide experimental validation of in silico predicted functions for three HP-encoding genes (yhdN, yeaC and ydgH) as proof of concept, by analyzing growth patterns of deletion mutants compared to the wild type, as well as their transcriptional responses to specific conditions. This study demonstrates that the use of Big Omics Data in combination with Artificial Intelligence and experimental controls is a powerful approach to illuminate functional dark matter.
Keywords: Artificial intelligence; Big omics data; Functional annotation of proteins; Functional dark matter; Independent Component Analysis (ICA).
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
