The concentration of single-stranded DNA-binding proteins is a critical factor in recombinase polymerase amplification (RPA), as revealed by insights from an open-source system
- PMID: 40821985
- PMCID: PMC12357552
- DOI: 10.7717/peerj.19758
The concentration of single-stranded DNA-binding proteins is a critical factor in recombinase polymerase amplification (RPA), as revealed by insights from an open-source system
Abstract
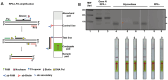
Recombinase polymerase amplification (RPA) facilitates rapid, exponential, isothermal nucleic acid amplification without the need for specialized equipment. Since its development in 2006, RPA has been widely applied to detect hundreds of RNA and DNA targets, spanning point-of-care diagnostics and agricultural uses. However, its reliance on pre-assembled commercial kits limits flexibility for customization. In this study, we introduce an open-source alternative to commercial RPA kits, utilizing purified, heterologously expressed proteins to circumvent the fixed molar ratios of proprietary systems. Our method incorporates enzymes from the bacteriophage T4 homologous recombination pathway-single-stranded binding protein (gp32), recombinase (UvsX), and mediator (UvsY)-along with Moloney murine leukemia virus (MMLV) reverse transcriptase with enhanced thermal stability, and Bst and Bsu DNA polymerases. We assessed the impact of buffer composition, reagent concentrations, and reaction temperature using synthetic SARS-CoV-2 genes. Notably, gp32 concentration and buffer composition emerged as critical factors in optimizing RPA performance. Using this tailored system, we demonstrated successful detection of the SARS-CoV-2 N gene on lateral flow devices (LFDs) with cDNA from eight clinical samples, achieving results consistent with RT-PCR. This open-source RPA platform provides an adaptable and cost-effective alternative for researchers, enabling the exploration of diverse experimental conditions and offering a viable solution for those without access to commercial kits.
Keywords: Point of care; Recombinase polymerase amplification; SARS-COV2.
©2025 Cordoba-Andrade et al.
Conflict of interest statement
Rogerio R. Sotelo-Mundo is an Academic Editor for PeerJ.
Figures



References
-
- Alberts BM, Morris C, Mace D, Sinha N, Bittner M, Moran L. DNA synthesis and its regulation. In: Goulian M, Hanawalt P, Fox CF, editors. ICN-UCLA symposia on molecular and cellular biology. Menlo Park: W. A. Benjamin, Inc.; 1975. pp. 241–269.
-
- Bleuit JS, Xu H, Ma Y, Wang T, Liu J, Morrical SW. Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8298–8305. doi: 10.1073/pnas.131007498. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

