Bioinformatics analysis of lncRNA and mRNA differentially expressed in patients with cervical cancer
- PMID: 40822023
- PMCID: PMC12354555
- DOI: 10.3389/fbinf.2025.1605681
Bioinformatics analysis of lncRNA and mRNA differentially expressed in patients with cervical cancer
Abstract
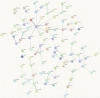
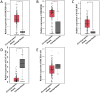
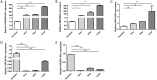
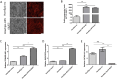
To verify the expression profile of long non-coding RNAs (lncRNAs) and mRNAs in cervical cancer, identify their clinical significance in HPV16-associated cervical cancer, and annotate the biological function of mRNAs. Three pairs of cancerous and paracancer tissues were selected in cervical squamous cell carcinoma (IB2 stage), high-throughput sequencing was utilized to determine the expression levels of lncRNAs and mRNAs. The detection results were validated by GEPIA database analysis and RT-qPCR. Functional annotations of differential mRNAs were conducted through Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and protein-protein interaction (PPI) network states. Furthermore, the association between antisense lncRNA and mRNA in cervical cancer was analyzed to predict the biological functions of lncRNA. Finally, recombinant lentivirus CV224-HPV16 E6/E7 was transfected into HcerEpic to establish a stable cell line with overexpressed HPV16 E6/E7, then differential lncRNAs were detected by RT-qPCR. Compared to paracancerous tissues, there were 3,608 lncRNAs significantly upregulated and 4,383 lncRNAs significantly downregulated in cervical cancer tissues (Fold change >2 and P < 0.05). Additionally, 3,666 mRNAs were significantly upregulated, while 2,220 mRNAs were significantly downregulated (Fold change >2 and P < 0.05). GO/KEGG enrichment analysis showed that differentially expressed mRNA played a significant role in cell cycle and cell senescence, and was related to signal pathways such as cAMP and MAPK, forming a complex network among the proteins encoded by these mRNAs. Further analysis indicated that the 20 antisense lncRNAs with the most remarkable differences might exert biological functions by influencing their corresponding mRNAs. The results of RT-qPCR revealed that CDKN2B-AS1, HAGLROS and GATA6-AS1 were potentially regulated by HPV16 E6/E7, which were in accordance with those obtained from chip detection. In this study, differentially expressed lncRNAs associated with HPV16 infection were screened and explored their transcriptional molecular functions and biological pathways, providing a molecular basis for predicting diagnostic markers of cervical cancer.
Keywords: HPV16 E6/E7; cervical cancer; high-throughput sequencing; lncRNA; mRNA.
Copyright © 2025 An, Huang, Yu, Tang, Wang, Chen, Zhang, Huang, Rao, Hu and Zha.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Identifying pyroptosis- and inflammation-related genes in spinal cord injury based on bioinformatics analysis.Sci Rep. 2025 Jul 14;15(1):25424. doi: 10.1038/s41598-025-10541-w. Sci Rep. 2025. PMID: 40659785 Free PMC article.
-
Dysregulated lncRNAs regulate human umbilical cord mesenchymal stem cell differentiation into insulin-producing cells by forming a regulatory network with mRNAs.Stem Cell Res Ther. 2024 Jan 25;15(1):22. doi: 10.1186/s13287-023-03572-5. Stem Cell Res Ther. 2024. PMID: 38273351 Free PMC article.
-
Screening and validation of long non-coding RNAs associated with colorectal cancer based on random forest and LASSO regression algorithm.Discov Oncol. 2025 Jul 1;16(1):1217. doi: 10.1007/s12672-025-03048-3. Discov Oncol. 2025. PMID: 40591188 Free PMC article.
-
Prophylactic vaccination against human papillomaviruses to prevent cervical cancer and its precursors.Cochrane Database Syst Rev. 2018 May 9;5(5):CD009069. doi: 10.1002/14651858.CD009069.pub3. Cochrane Database Syst Rev. 2018. PMID: 29740819 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
LinkOut - more resources
Full Text Sources

