Phylogeny-guided discovery of a promiscuous P450 macrocyclase for the production of diverse atropopeptides
- PMID: 40822110
- PMCID: PMC12352619
- DOI: 10.1039/d5sc03525b
Phylogeny-guided discovery of a promiscuous P450 macrocyclase for the production of diverse atropopeptides
Abstract
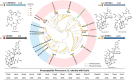
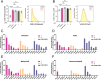
Cyclic peptides exhibit diverse bioactivities and are distinguished by their enhanced cell permeability, improved proteolytic stability, and increased binding affinity due to their conformational rigidity. Despite significant advancements in peptide synthesis, the production of complex cyclic peptides remains a challenge. Nature has evolved diverse strategies for peptide cyclization, with an ever-growing repertoire of characterized cyclases involved in the biosynthesis of ribosomally synthesized and post-translationally modified peptides (RiPPs). These enzymes convert linear precursor peptides into complex (poly-)cyclic structures. The discovery of the atropopeptides has significantly expanded the chemical diversity of RiPPs with unique (poly-)cyclic structures. In this study, we employed a phylogeny-guided approach to identify a substrate-promiscuous cytochrome P450 macrocyclase that catalyzes the formation of cyclic peptides through atropospecific C-N or C-C bond formation between aromatic amino acid side chains. Combinatorial biosynthetic studies revealed that ScaB encoded in the scabrirubin biosynthetic gene cluster efficiently cyclizes a wide range of atropopeptide precursor peptides. Furthermore, extensive site-directed mutagenesis studies of the tetrapeptide core sequence further expanded the diversity of atropopeptides. Notably, three tested atropopeptides show antiviral activity and one of the non-natural atropopeptides displays anti-inflammatory activity. Our findings establish a broadly substrate-tolerant atropopeptide-modifying P450 as a versatile biocatalyst for the synthesis of bioactive, biaryl-bridged macrocyclic peptides.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Similar articles
-
Plant peptides - redefining an area of ribosomally synthesized and post-translationally modified peptides.Nat Prod Rep. 2024 Jul 17;41(7):1020-1059. doi: 10.1039/d3np00042g. Nat Prod Rep. 2024. PMID: 38411572 Free PMC article. Review.
-
Molecular Basis for Peptide Nitration by a Novel Cytochrome P450 Enzyme in RiPP Biosynthesis.ACS Catal. 2025 Jun 3;15(12):10391-10404. doi: 10.1021/acscatal.5c01932. eCollection 2025 Jun 20. ACS Catal. 2025. PMID: 40568218 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Genome mining identifies a diversity of natural product biosynthetic capacity in human respiratory Corynebacterium strains.mSphere. 2025 Jun 25;10(6):e0025825. doi: 10.1128/msphere.00258-25. Epub 2025 May 21. mSphere. 2025. PMID: 40396729 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
-
- Kwon Y. U. Kodadek T. Quantitative comparison of the relative cell permeability of cyclic and linear peptides. Chem. Biol. 2007;14:671–677. - PubMed
-
- Baeriswyl V. Heinis C. Phage selection of cyclic peptide antagonists with increased stability toward intestinal proteases. Protein Eng. Des. Sel. 2013;26:81–89. - PubMed
-
- Molhoek E. M. van Dijk A. Veldhuizen E. J. A. Haagsman H. P. Bikker F. J. Improved proteolytic stability of chicken cathelicidin-2 derived peptides by D-amino acid substitutions and cyclization. Peptides. 2011;32:875–880. - PubMed
-
- Yao J. F. Yang H. Zhao Y. Z. Xue M. Metabolism of Peptide Drugs and Strategies to Improve their Metabolic Stability. Curr. Drug Metab. 2018;19:892–901. - PubMed
LinkOut - more resources
Full Text Sources

