C-BIOPRED severe asthma clinical phenotypes: link to complement and coagulation pathways and galectin 10
- PMID: 40822235
- PMCID: PMC12351390
- DOI: 10.1183/23120541.01016-2024
C-BIOPRED severe asthma clinical phenotypes: link to complement and coagulation pathways and galectin 10
Abstract
Background: Severe asthma is a heterogeneous airway inflammatory disease presenting with varying clinicophysiological characteristics and response to treatments. The objectives of the present study were to determine the clinical phenotypes of the Chinese C-BIOPRED cohort and their link to the sputum proteome.
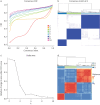
Methods: Partition-around-medoids clustering was applied to a training set of 362 nonsmoking, smoking or ex-smoking severe asthma patients, and nonsmoking mild-moderate asthma patients using eight clinicophysiological variables, with validation performed in the remaining 181.
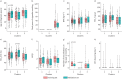
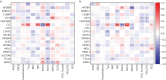
Results: Three stable clusters were defined, with Cluster T1 composed of predominantly female patients with severe nonsmoking asthma experiencing frequent exacerbations with moderate airflow obstruction, and Cluster T3 of elderly male patients with smoking/ex-smoking late-onset severe asthma and severe airflow obstruction and a moderate number of exacerbations. Cluster T2 was composed of nonsmokers with a mild-moderate airflow obstruction and no previous exacerbations. Validation clusters (V1, V2 and V3) were similar to the training set clusters. Differentially expressed proteins in sputum supernatants measured by liquid chromatography with tandem mass spectrometry pointed to differences in the complement and coagulation cascade pathway between Cluster 1 (T1 and V1) and Cluster 3 (T3 and V3), as well as between Cluster 2 (T2 and V2) and Cluster 3. Galectin 10 was upregulated in Cluster 1 compared with Cluster 2, and correlated with exacerbations, fractional exhaled nitric oxide, blood and sputum eosinophil count and oral corticosteroid dose in Cluster 1.
Conclusion: The clinical clusters were differentiated by smoking status, degree of airflow obstruction and exacerbation history, and by sputum complement and coagulation pathways, and galectin 10 levels.
Copyright ©The authors 2025.
Conflict of interest statement
Conflict of interest: K.F. Chung reports personal fees from attending advisory board meetings with GSK, AZ, Novartis, Roche, Merck, Trevi, Rickett-Beckinson, Nocion and Shionogi; is a scientific adviser to The Clean Breathing Institute supported by Haleon; reports personal fees for speaking at meetings supported by GSK, Sanofi, Novartis and AZ; and, through his institution, has received research funding from Merck and GSK. The other authors have no relevant conflicts of interest.
Figures



References
LinkOut - more resources
Full Text Sources
