Meropenem/vaborbactam activity against carbapenem-resistant Klebsiella pneumoniae from catheter-related bloodstream infections
- PMID: 40822579
- PMCID: PMC12350291
- DOI: 10.3389/fcimb.2025.1616353
Meropenem/vaborbactam activity against carbapenem-resistant Klebsiella pneumoniae from catheter-related bloodstream infections
Abstract
Introduction: Carbapenem-resistant Klebsiella pneumoniae (CRKP) poses a significant threat in oncology settings due to its multidrug resistance and ability to form biofilms on indwelling medical devices.
Methods: This study investigated the in vitro and in vivo activity of meropenem/vaborbactam (MEV) against two CRKP isolates recovered from catheter-related bloodstream infections in patients undergoing orthopedic oncologic surgery.
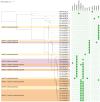
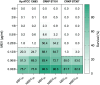
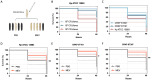
Results: Whole-genome sequencing identified the isolates as ST101 and ST307, harboring resistance determinants including blaKPC-3 and blaOXA-1 , distributed across IncFII and IncFIB plasmid replicons. Both isolates exhibited extensive resistance to β-lactams, aminoglycosides, and fluoroquinolones but remained susceptible to MEV. Phenotypic assays revealed enhanced biofilm formation and metabolic activity compared to the reference strain Kp ATCC 13883 in the absence of hypervirulence-associated genes. MEV demonstrated bactericidal activity against both planktonic and biofilm-associated cells, with minimum bactericidal concentration (MBC90) and minimum biofilm eradication concentration (MBEC90) values of 0.5/8 μg/ml for CRKP ST101, 0.12/8 μg/ml for CRKP ST307, and 0.25/8 μg/ml for the Kp ATCC 13883 strain. In the Galleria mellonella infection model, MEV significantly improved larval survival following the CRKP challenge.
Discussion: These findings demonstrate that MEV exhibits activity against planktonic and biofilm-associated CRKP cells and highlight the need for further investigation in managing catheter-related bloodstream infections caused by multidrug-resistant K. pneumoniae.
Keywords: biofilm-associated infections; carbapenem-resistant Klebsiella pneumoniae; catheter-related bloodstream infections; meropenem/vaborbactam; multidrug-resistant plasmid; surgical site infection.
Copyright © 2025 Sivori, Francalancia, Truglio, Cavallo, Pronesti, Fabrizio, Celesti, Cazzani, Furzi, Pimpinelli and Di Domenico.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer MM declared a shared affiliation with the author GF to the handling editor at the time of review. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



References
-
- Alcock B. P., Huynh W., Chalil R., Smith K. W., Raphenya A. R., Wlodarski M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920, PMID: - DOI - PMC - PubMed
-
- Benthall G., Touzel R. E., Hind C. K., Titball R. W., Sutton J. M., Thomas R. J., et al. (2015). Evaluation of antibiotic efficacy against infections caused by planktonic or biofilm cultures of Pseudomonas aeruginosa and Klebsiella pneumoniae in Galleria mellonella. Int. J. Antimicrobial. Agents 46, 538–545. doi: 10.1016/j.ijantimicag.2015.07.014, PMID: - DOI - PubMed
-
- Bianco G., Boattini M., Lupo L., Ambretti S., Greco R., Degl’Innocenti L., et al. (2025). In vitro activity and genomic characterization of KPC-producing Klebsiella pneumoniae clinical blood culture isolates resistant to ceftazidime/avibactam, meropenem/vaborbactam, imipenem/relebactam: an Italian nationwide multicentre observational study, (2022–23). J. Antimicrobial. Chemother. 80, 583–592. doi: 10.1093/jac/dkae450, PMID: - DOI - PubMed
-
- Bonura C., Giuffrè M., Aleo A., Fasciana T., Di Bernardo F., Stampone T., et al. (2015). An Update of the Evolving Epidemic of blaKPC Carrying Klebsiella pneumoniae in Sicily, Italy 2014: emergence of multiple non-ST258 clones. PloS One 10, e0132936. doi: 10.1371/journal.pone.0132936, PMID: - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

