Genetic characterization of Lynch syndrome germline variants in a LATAM cohort using a customized NGS gene panel
- PMID: 40823072
- PMCID: PMC12353692
- DOI: 10.3389/fonc.2025.1589765
Genetic characterization of Lynch syndrome germline variants in a LATAM cohort using a customized NGS gene panel
Abstract
Introduction: Lynch Syndrome accounts for 1-7% of all colorectal cancers and is caused by germline mutations in DNA mismatch repair (MMR) genes. Timely molecular diagnosis is crucial for effective genetic counseling and management. Among understudied Latin American populations, Uruguay's genetic admixture provides an opportunity to identify novel Lynch Syndrome related variants.
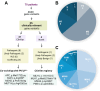
Methods: This study analyzed 70 unrelated Uruguayan colorectal cancer patients meeting Lynch Syndrome clinical criteria to identify carriers of pathogenic variants. A customized Next-Generation Sequencing (NGS) panel was developed and sequenced on the Ion Torrent platform to analyze nine genes: MLH1, MSH2, MSH6, EPCAM, FAN1, MUTYH, PMS1, PMS2, and APC. Copy number variations and large EPCAM deletions are not detected by the assay. Gene variants were prioritized based on allelic frequency, in silico predictions, pathogenicity records, and ACMG guidelines. The performance of this custom NGS panel was evaluated for in-house applications, and its limitations were thoroughly assessed.
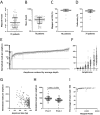
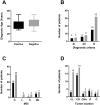
Results and discussion: The custom NGS panel demonstrated effectiveness for scalable in-house testing despite minor disclosed sequence coverage limitations. Pathogenic and likely pathogenic variants were identified in 25 patients, including four novel Lynch Syndrome-associated variants. In four patients, a rare ambiguously classified gene variant co-occurs with a known pathogenic variant in another gene. The mutation profile correlated with clinical parameters such as age of diagnosis, diagnosis criteria, tumor location, and microsatellite instability (MSI).
Conclusion: This is the most comprehensive genetic study to date on a Uruguayan Lynch syndrome cohort. The mutational landscape aligns with findings in other populations while highlighting novel variants of clinical relevance. These findings highlight the value of customized panels for improving genetic screening in small-scale healthcare facilities.
Keywords: LATAM; Lynch Syndrome; NGS; colorectal cancer; germline cancer predisposition; novel variants.
Copyright © 2025 Mathó, Chávez, Fort, Della Valle, Neffa, Sotelo-Silveira, Artagaveytia and Duhagon.
Conflict of interest statement
CM, SC, RSF, JS-S, and MD are researchers of the Sistema Nacional de Investigadores SNI of the Agencia Nacional de Investigación e Innovación ANII, Uruguay. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- National Comprehensive Cancer Network Guidelines . National comprehensive cancer network guidelines. In: NCCN clinical practice guidelines in oncology (NCCN guidelines®) (2023). Available online at: https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

