Impact of genetic variants linked to liver fat and liver volume on MRI-mapped body composition
- PMID: 40823175
- PMCID: PMC12355076
- DOI: 10.1016/j.jhepr.2025.101468
Impact of genetic variants linked to liver fat and liver volume on MRI-mapped body composition
Abstract
Background & aims: A quarter of the world population is estimated to have metabolic dysfunction-associated steatotic liver disease. Here, we aim to understand the impact of liver trait-associated genetic variants on fat content and tissue volume across organs and body compartments and on a large set of biomarkers.
Methods: Genome-wide association analyses were performed on liver fat and liver volume estimated with magnetic resonance imaging in up to 27,243 unrelated European participants from the UK Biobank. Identified variants were assessed for associations with fat fraction and tissue volume in >2 million 'Imiomics' image elements in 22,261 individuals and with circulating biomarkers in 310,224 individuals.
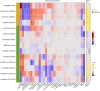
Results: We confirmed four liver fat and nine liver volume previously reported genetic variants (p values <5 × 10-8). We further found evidence suggestive of a novel liver volume locus, ADH4, where each additional T allele increased liver volume by 0.05 SD (SE = 0.01, p value = 3.3 × 10-8). The Imiomics analyses showed that liver fat-increasing variants were specifically associated with fat fraction of the liver tissue (p values <2.8 × 10-3) and with higher inflammation, liver and renal injury biomarkers, and lower lipid levels. Associations of liver volume variants with fat content, tissue volume, and biomarkers were more heterogeneous, for example the liver volume-increasing alleles at CENPW and PPP1R3B were associated with higher skeletal muscle volumes and were more pronounced in men, whereas the GCKR variant was negatively associated with lower skeletal muscle volumes in women (p values <2.8 × 10-3).
Conclusions: Liver fat-increasing variants were mostly linked to fat fraction of the liver and were positively associated with some adverse metabolic biomarkers and negatively with lipids. In contrast, liver volume-associated variants showed a less consistent pattern across organs and biomarkers.
Impact and implications: Liver fat and liver volume are common metabolic traits with a strong genetic component, yet the extent to which they exert organ-specific vs. systemic effects remains poorly defined. By integrating genome-wide association analyses and high-resolution neck-to-knee magnetic resonance imaging data through the Imiomics framework, this study reveals distinct genetic architectures for liver fat and liver volume, including sex-specific effects. These findings provide new insights into the biological, organ-level, tissue-specific, and systemic characteristics of steatotic liver disease and its genetic determinants. The results may inform the development of precision imaging genetic approaches, biomarker discovery, and stratified risk assessment strategies, while reinforcing the importance of incorporating sex-specific analyses in future research and clinical applications.
Keywords: Chronic liver disease; Genetic variation; Metabolic disease; Metabolic dysfunction-associated steatotic liver disease.
© 2025 The Authors.
Conflict of interest statement
JK and HA are co-founders, co-owners of, and part-time employees of Antaros Medical AB, BioVenture Hub, Mölndal, Sweden. JCC receives a salary from Sirona AB. Her contribution to this manuscript was performed prior to commencing employment. GDC was affiliated with the University of Copenhagen during the conduct of this research. He is currently employed at Novo Nordisk. This work was completed before his employment and was not funded or influenced by Novo Nordisk. Other co-authors reported no potential conflicts of interest relevant to this article. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures



References
-
- Byrne C.D., Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62:S47–S64. - PubMed
-
- Ahmed A., Cule M., Bell J.D., et al. Differing genetic variants associated with liver fat and their contrasting relationships with cardiovascular diseases and cancer. J Hepatol. 2024;81:921–929. - PubMed
-
- Targher G., Byrne C.D., Tilg H. MASLD: a systemic metabolic disorder with cardiovascular and malignant complications. Gut. 2024;73:691–702. - PubMed
LinkOut - more resources
Full Text Sources

