Development and validation of an interpretable machine learning model for retrospective identification of suspected infection for sepsis surveillance: a multicentre cohort study
- PMID: 40823498
- PMCID: PMC12355416
- DOI: 10.1016/j.eclinm.2025.103401
Development and validation of an interpretable machine learning model for retrospective identification of suspected infection for sepsis surveillance: a multicentre cohort study
Abstract
Background: How to identify suspected infection for sepsis surveillance purposes remains a well-recognised challenge. This study aimed to operationalise suspected infection for sepsis surveillance by developing an interpretable machine learning (ML) model for retrospective identification of patients with sepsis.
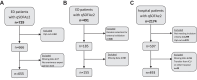
Methods: This multicentre cohort and machine learning study was conducted in two Dutch tertiary care hospitals. Adult patients with a quick Sequential Organ Failure assessment (qSOFA) ≥2 were included. Exclusion criteria included admission to the intensive care unit, transfer to or from another hospital, or patient refusal to reuse data. Cohort one consisted of patients admitted to the Emergency Department (ED) of hospital A between 01/01/2019 and 12/31/2019, to investigate community-onset sepsis. An external validation cohort of ED patients was obtained from hospital B between 01/01/2021 and 06/03/2022. Cohort two included hospitalised patients from hospital A between 01/01/2021 and 06/01/2022, to investigate hospital-onset sepsis. Objective data were extracted from electronic health records. Seven ML methods, including gradient boosting, random forest, logistic regression, decision trees, support vector machines, K nearest neighbours and stochastic gradient descent, were trained to identify sepsis with manual chart review as reference standard. The F1 score (harmonic mean of precision and recall), sensitivity and specificity were used as evaluation metrics. The best performing ML method was compared with other commonly used suspected infection proxies, including the Sepsis-3 definition, an adapted Adult Sepsis Event (ASE) definition and International Classification of Diseases (ICD) codes.
Findings: In the ED cohort, 655 patients were included (male: 355 (54.2%), female: 300 (45.8%)) and 240 (36.6%) had sepsis. For community-onset sepsis, gradient boosting performed best with an F1 score of 85.9%, a sensitivity of 91.1% (95%-CI 83.4-95.4%) and a specificity of 89.0% (95%-CI 83.4-92.8%). Most model features reflected either the inflammatory response (CRP, body temperature) or actions taken when an infection is suspected (antibiotic administration, microbial culture). In the external validation cohort, 185 patients were included (male: 94 (50.8%), female: 91 (49.2%)) and 54 (29.2%) had sepsis. External validation yielded an F1 score of 85.7%, a sensitivity of 87.5% (95%-CI 75.3-94.1%) and a specificity of 92.5% (95%-CI 85.9-96.2%). The gradient boosting model outperformed other commonly used proxies for suspected infection in terms of sensitivity, achieving 91.1% (95% CI: 83.4-95.4%), compared to Sepsis-3 with 78.9% (95% CI: 69.4-86.0%), the adapted ASE with 85.6% (95% CI: 76.8-91.4%), and ICD codes with 33.3% (95% CI: 24.5-43.6%). In the hospitalised cohort, 493 patients were included (male: 265 (53.8%), female: 228 (46.2%)) and 129 (26.2%) had sepsis. For hospital-onset sepsis, logistic regression had the highest F1 score (52.2%). Sensitivity was 58.1% (95%-CI 40.6-75.5%) and specificity was 82.9% (95%-CI 76.0-89.8%).
Interpretation: ED patients meeting ≥2 qSOFA criteria can be accurately classified as having suspected infection or not by a gradient boosting algorithm, outperforming common suspected infection definitions for sepsis surveillance. Including the inflammatory response in the suspected infection surveillance definition may enhance the accuracy and objectivity of sepsis surveillance. Future research is needed to validate the algorithm using other organ dysfunction criteria and in international settings.
Funding: None.
Keywords: Machine learning; Sepsis; Sepsis surveillance; Suspected infection.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Yu S.C., Betthauser K.D., Gupta A., et al. Comparison of sepsis definitions as automated criteria. Crit Care Med. 2021;49(4):e433–e443. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

