Cardiovascular diseases in the elderly: possibilities for modulating autophagy using non-coding RNAs
- PMID: 40823532
- PMCID: PMC12350270
- DOI: 10.3389/fcell.2025.1520850
Cardiovascular diseases in the elderly: possibilities for modulating autophagy using non-coding RNAs
Abstract
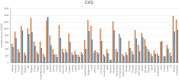
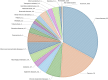
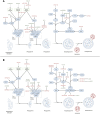
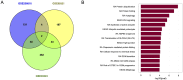
Autophagy is a crucial mechanism implicated in both aging and cardiovascular disease, which are two closely interconnected conditions. Modulation of autophagy is expected to have profound impacts on cellular aging and maintenance of cardiovascular functions under physiological or pathological conditions. Consequently, modulation of autophagy could be an effective strategy for counteracting age-induced vascular and cardiac remodelling as well as alleviating cardiovascular disease. The present review comprehensively elucidates the multifaceted impacts of autophagy on aging of the cardiovascular system. We comprehensively analyse both vascular and cardiac tissues, including vascular and cardiac malignancies, in distinct contexts. We also emphasize the significance of non-coding RNAs (ncRNAs) in the epigenetic regulation of gene expression and their roles as biomarkers of cardiovascular pathologies while maintaining clear distinctions between the vascular and cardiac tissues. Preclinical and clinical models are described herein to highlight the importance of ncRNAs in disease treatment by considering their involvement in the modulation of autophagy within the cardiocirculatory system. Finally, we conducted a comprehensive meta-analysis of transcriptomic data to underscore the paramount importance of autophagy while demonstrating it as a process that is frequently dysregulated in both cardiac and vascular cells under pathological conditions. The findings presented herein emphasize the importance of investigating novel strategies for modulating autophagy as a potential therapeutic approach to the management of age-related cardiovascular disorders.
Keywords: aging; autophagy; cardiovascular disease; meta-analysis; non-coding RNAs.
Copyright © 2025 Scalabrin and Cagnin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
Figures
References
-
- Aghagolzadeh P., Plaisance I., Bernasconi R., Treibel T. A., Pulido Quetglas C., Wyss T., et al. (2023). Assessment of the cardiac noncoding transcriptome by single-cell RNA sequencing identifies FIXER, a conserved profibrogenic long noncoding RNA. Circulation 148, 778–797. 10.1161/CIRCULATIONAHA.122.062601 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

