ONE-STEP tagging: a versatile method for rapid site-specific integration by simultaneous reagent delivery
- PMID: 40823809
- PMCID: PMC12359036
- DOI: 10.1093/nar/gkaf809
ONE-STEP tagging: a versatile method for rapid site-specific integration by simultaneous reagent delivery
Abstract
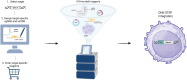
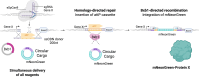
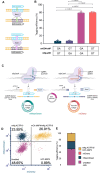
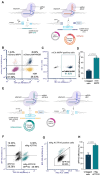
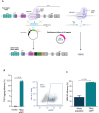
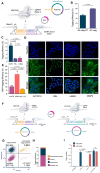
We present a novel, versatile genome editing method termed ONE-STEP tagging, which combines CRISPR-Cas9-mediated targeting with Bxb1 integrase-based site-specific integration for efficient, precise, and scalable protein tagging. Applied in human-induced pluripotent stem cells (hiPSCs), cancer cells and primary T cells, this system enables rapid generation of endogenously tagged proteins. By enhancing the nuclear localization signal of the catalytically superior eeBxb1 integrase and co-delivering a DNA-PK inhibitor, we achieved up to ∼90% integration efficiency at the ACTR10 locus in hiPSCs. ONE-STEP tagging is robust across loci and cell types and supports large DNA cargo integration, with efficiencies reaching 16.6% for a 14.4 kb construct. The method also enables multiplexed tagging of multiple proteins within the same cell and simultaneous CRISPR-based editing at secondary loci, such as gene knockouts or homology-directed repair. Importantly, we demonstrate successful application in primary T cells by targeting the T cell receptor locus while simultaneously knocking out B2M, a key step towards generating immune-evasive, off-the-shelf chimeric antigen receptor T cells. Additionally, we introduce a dual-cassette version of the method compatible with universal donor plasmids, allowing use of entirely off-the-shelf reagents. Together, these advances establish ONE-STEP tagging as a powerful tool for both basic and therapeutic genome engineering.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
A.R.B. is a founder of and consultant for Ensocell therapeutics. A.R.B., V.M., and T.B. are inventors on patent US20250002943A1, EP4408993A1, and WO2023052774A1 related to this work and another application is in process.
Figures

References
-
- Li T, Yang Y, Qi H et al. CRISPR–Cas9 therapeutics: progress and prospects. Nature. 2023; 8:113–7. 10.1038/s41392-023-01309-7. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

