Controlling gene expression through five zinc finger domains of ZNF18
- PMID: 40823929
- PMCID: PMC12359201
- DOI: 10.1002/pro.70278
Controlling gene expression through five zinc finger domains of ZNF18
Abstract
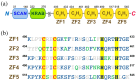
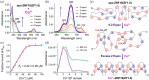
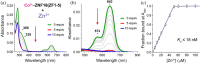
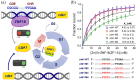
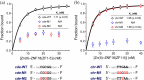
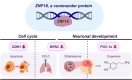
Zinc finger (ZF) proteins are the most abundant transcription factors in vertebrates, and they regulate gene expression through interactions with cis-acting elements. ZF domains selectively recognize specific sequences to accelerate or repress target genes. Zinc finger protein 18 (ZNF18) contains five CX2CX12HX3H-type ZFs at the C-terminus, which are expressed in the brain and other organs of the biological system. Bioinformatic study proposed that cyclin-dependent kinase 1 (CDK1) is in the signaling cascade of ZNF18; although experimental evidence has not yet been reported. In this study, we expressed and purified ZNF18(ZF1-5), five ZF domains from ZNF18, and investigated metal binding specificity and promoter interactions. ZNF18(ZF1-5) has specific coordination to Zn2+ (Kd ≤ 18 nM) compared with other xenobiotic metal ions, including Co2+, Fe2+, and Fe3+, with 98.5% of reduced ZF domains after purification. This significantly active ZF can be one of the major reasons for tight coordination affinity. CDK1 rescued the arrested cell cycle induced by DNA damage, resulting in tumorigenesis. Zn2+-ZNF18(ZF1-5) specifically binds to cis-acting elements of cdk1 (Kd = 4.63 ± 0.07 nM), mediated by a cell cycle-dependent element (cde, 5'-CGCGG) and a cell cycle gene homology region (chr, 5'-TTGAA). The ZNF18 superfamily was expressed in the brain for the regulation of neuronal development and cell differentiation. Zn2+-ZNF18(ZF1-5) interacted with promoters in the insulin response sequence (IRS) for inhibition of dopamine secretion and cis-acting element of brain-2 (BRN2), which controlled astrocyte and cancer development. These results provide the first evidence that ZNF18(ZF1-5) regulates the cell cycle and neuronal development through transcriptional regulation.
Keywords: apoptosis; astrocyte; cell cycle regulation; classic zinc fingers; cyclin‐dependent kinase 1; transcription factor; zinc finger protein 18 (ZNF18).
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors have no conflicts of interest requiring disclosure.
Figures



References
-
- Berg JM, Godwin HA. Lessons from zinc‐binding peptides. Annual Review of Biophysics and Biomolecular Structure. 1997;26:357–371. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

