Identifying microbial functional guilds performing cryptic organotrophic and lithotrophic redox cycles in anaerobic granular biofilms
- PMID: 40824903
- PMCID: PMC12360541
- DOI: 10.1371/journal.pone.0330380
Identifying microbial functional guilds performing cryptic organotrophic and lithotrophic redox cycles in anaerobic granular biofilms
Abstract
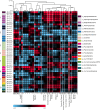
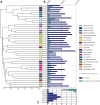
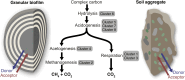
Granular biofilms used in anaerobic digester systems contain diverse microbial populations that interact to hydrolyze organic matter and produce methane within controlled environments. Prior research investigated the feasibility of utilizing granular biofilms obtained from an anaerobic digester to remove nitrate without the addition of exogenous electron donors. These granules possessed a unique structure of alternating light and dark iron sulfide and pyrite rich layers that potentially served as both an electron source and sink, linking carbon, nitrogen, sulfur, and iron cycles. To characterize the functional roles of diverse microbial populations enriched within these layered biofilms, we analyzed metagenomes obtained from three different granules. Comparisons between the functional gene content of forty metagenome assembled genomes (MAGs) identified phylogenetically cohesive functional guilds. Each of these functional MAG clusters was assigned to specific steps in anaerobic digestion (hydrolysis, acidogenesis, acetogenesis, and methanogenesis) and anaerobic respiration (denitrification and sulfate reduction). Comparisons with metagenomes derived from a variety of natural and engineered ecosystems confirmed that the enriched denitrifying bacteria were similar to populations typically found in wetlands and biological nitrogen removal systems. Analysis of read alignments to individual genes within the forty MAGs identified conserved genomic features that were representative of the functions that distinguished functional guilds. Overall, this research illustrates the utility of functional based classification of microorganisms for characterizing ecosystem functions and highlights the potential application of engineered ecosystems to serve as experimental models for complex natural ecosystems.
Copyright: © 2025 Flinkstrom et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- McHugh S, O’Reilly C, Mahony T, Colleran E, O’Flaherty V. Anaerobic Granular Sludge Bioreactor Technology. Rev Environ Sci Technol. 2003;2:225–45.
-
- Skiadas IV, Gavala HN, Schmidt JE, Ahring BK. Anaerobic Granular Sludge and Biofilm Reactors. In: Ahring BK, Ahring BK, Angelidaki I, Dolfing J, EUegaard L, Gavala HN, editors. Biomethanation II. Berlin, Heidelberg: Springer Berlin Heidelberg. 2003. p. 35–67. - PubMed
-
- O’Flaherty V, Collins G, Mahony T. The Microbiology and Biochemistry of Anaerobic Bioreactors with Relevance to Domestic Sewage Treatment. Rev Environ Sci Biotechnol. 2006;5(1):39–55. doi: 10.1007/s11157-005-5478-8 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

