Investigation of mobile genetic elements and their association with antibiotic resistance genes in clinical pathogens worldwide
- PMID: 40824964
- PMCID: PMC12360581
- DOI: 10.1371/journal.pone.0330304
Investigation of mobile genetic elements and their association with antibiotic resistance genes in clinical pathogens worldwide
Abstract
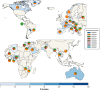
Objectives: Antimicrobial-resistant bacteria are a major global health threat. Mobile genetic elements (MGEs) have been crucial for spreading resistance to new bacterial species, including human pathogens. Understanding how MGEs promote resistance could be essential for prevention. Here we present an investigation of MGEs and their association with resistance genes in pathogenic bacteria collected from 59 diagnostic units during 2020, representing a snapshot of clinical infections from 35 counties worldwide.
Methods: We analysed 3,095 whole-genome sequenced clinical bacterial isolates from over 100 species to study the relationship between resistance genes and MGEs. The mobiliome of Staphylococcus aureus, Enterococcus faecalis, Escherichia coli, and Klebsiella pneumoniae were further examined for geographic differences, as these species were prevalent in all countries. Genes potentially mobilized by MGEs were identified by finding DNA segments containing MGEs and ARGs preserved in multiple species. Network analysis was used to investigate potential MGE interactions, host range, and transmission pathways.
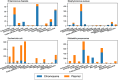
Results: The prevalence and diversity of MGEs and resistance genes varied among species, with E. coli and S. aureus carrying more diverse elements. MGE composition differed between bacterial lineages, indicating strong vertical inheritance. 102 MGEs associated with resistance were found in multiple species, and four of these elements seemed to be highly transmissible as they were found in different phyla. We identified 21 genomic regions containing resistance genes potentially mobilized by MGEs, highlighting their importance in transmitting genes to clinically significant bacteria.
Conclusion: Resistance genes are spread through various MGEs, including plasmids and transposons. Our findings suggest that multiple factors influence MGE prevalence and their transposability, thereby shaping the MGE population and transmission pathways. Some MGEs have a wider host range, which could make them more important for mobilizing genes. We also identified 103 resistance genes potentially mobilised by MGEs, which could increase their transmissibility to unrelated bacteria.
Copyright: © 2025 Johansson et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- World Health Organization. Antimicrobial resistance: Global Report on Surveillance. 2014 [cited 4 May 2024]. Available from: https://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf
-
- World Health Organization. Global Priority List of Antibiotic-Resistance Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017 [cited 9 Jun 2023]. Available from: https://remed.org/wp-content/uploads/2017/03/lobal-priority-list-of-anti...

