Dysregulated SASS6 expression promotes increased ciliogenesis and cell invasion phenotypes
- PMID: 40825584
- PMCID: PMC12361644
- DOI: 10.26508/lsa.202402820
Dysregulated SASS6 expression promotes increased ciliogenesis and cell invasion phenotypes
Abstract
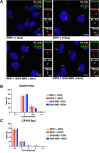
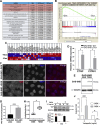
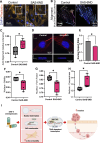
Centriole and/or cilium defects are characteristic of cancer cells and have been linked to cancer cell invasion. However, the mechanistic bases of this regulation remain incompletely understood. Spindle assembly abnormal protein 6 homolog (SAS-6) is essential for centriole biogenesis and cilium formation. SAS-6 levels decrease at the end of mitosis and G1, resulting from APCCdh1-targeted degradation. To examine the biological consequences of unrestrained SAS-6 expression, we used a nondegradable SAS-6 mutant (SAS-6ND). This led to an increase in ciliation and cell invasion and caused an up-regulation of the YAP/TAZ pathway. SAS-6ND expression resulted in cell morphology changes, nuclear deformation, and YAP translocation to the nucleus, resulting in increased TEAD-dependent transcription. SAS-6-mediated invasion was prevented by YAP down-regulation or by blocking ciliogenesis. Similarly, down-regulation of SAS-6 in DMS273, a highly invasive and highly ciliated lung cancer cell line that overexpresses SAS-6, completely blocked cell invasion and depleted YAP protein levels. Thus, our data provide evidence for a defined role of SAS-6 in cell invasion through the activation of the YAP/TAZ pathway.
© 2025 Hargreaves et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures












References
-
- Bergwell M, Smith A, Smith E, Dierlam C, Duran R, Haastrup E, Napier-Jameson R, Seidel R, Potter W, Norris A, et al. (2023) A primary microcephaly-associated sas-6 mutation perturbs centrosome duplication, dendrite morphogenesis, and ciliogenesis in Caenorhabditis elegans. Genetics 224: iyad105. 10.1093/genetics/iyad105 - DOI - PMC - PubMed
-
- Brodowska K, Al-Moujahed A, Marmalidou A, Meyer Zu Horste M, Cichy J, Miller JW, Gragoudas E, Vavvas DG (2014) The clinically used photosensitizer Verteporfin (VP) inhibits YAP-TEAD and human retinoblastoma cell growth in vitro without light activation. Exp Eye Res 124: 67–73. 10.1016/j.exer.2014.04.011 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
