Dissecting pOXA-48 fitness effects in clinical Enterobacterales using plasmid-wide CRISPRi screens
- PMID: 40825783
- PMCID: PMC12361523
- DOI: 10.1038/s41467-025-63082-1
Dissecting pOXA-48 fitness effects in clinical Enterobacterales using plasmid-wide CRISPRi screens
Abstract
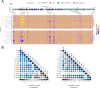
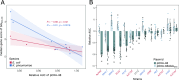
Conjugative plasmids are the main vehicle for the spread of antimicrobial resistance (AMR) genes in clinical bacteria. AMR plasmids allow bacteria to survive antibiotic treatments, but they also produce physiological alterations in their hosts that commonly translate into fitness costs. Despite the key role of plasmid-associated fitness effects in AMR evolution, their origin and molecular bases remain poorly understood. In this study, we introduce plasmid-wide CRISPR interference (CRISPRi) screens as a tool to dissect plasmid-associated fitness effects. We design and perform CRISPRi screens targeting the globally distributed carbapenem resistance plasmid pOXA-48 in 13 different multidrug resistant clinical Enterobacterales. Our results reveal that pOXA-48 gene-level effects are conserved across clinical strains, and expose the key role of the carbapenemase-encoding gene, blaOXA-48, as the main culprit for pOXA-48 fitness costs. Moreover, our results highlight the relevance of postsegregational killing systems in pOXA-48 vertical transmission, and uncover new genes implicated in pOXA-48 stability (pri, korC, DNDJGHEP_13 and 14 and H-NS). This study sheds new light on the biology and evolution of carbapenem resistant Enterobacterales and endorses CRISPRi screens as a powerful method for studying plasmid-mediated AMR.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Review on Antimicrobial Resistance. Antimicrobial Resistance - Tackling a Crisis for the Health and Wealth of Nations. (2014).
-
- Ho, C. S. et al. Antimicrobial resistance: a concise update. Lancet Microbe0, 100947 (2024). - PubMed
-
- World Bank. Drug-Resistant Infections: A Threat to Our Economic Future. (2017).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

