Noncanonical function of Pannexin1 promotes cellular senescence and renal fibrosis post-acute kidney injury
- PMID: 40825974
- PMCID: PMC12361471
- DOI: 10.1038/s41467-025-63152-4
Noncanonical function of Pannexin1 promotes cellular senescence and renal fibrosis post-acute kidney injury
Abstract
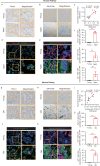
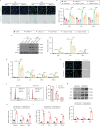
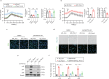
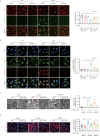
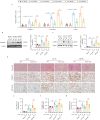
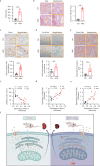
Acute kidney injury (AKI) can lead to chronic kidney disease (CKD), a transition driven by cellular senescence, a state of irreversible cell-cycle arrest. However, the molecular mechanisms promoting this pathological process remain unclear. Here we show that the channel protein Pannexin1 (Panx1) promotes this detrimental senescence and subsequent kidney fibrosis. We found that Panx1 functions in a noncanonical role as a calcium (Ca2+) leak channel within the endoplasmic reticulum (ER), a key intracellular calcium store. This Panx1-mediated leak occurs at contact sites between the ER and mitochondria, leading to mitochondrial calcium overload, dysfunction, and the generation of pro-senescence signals. Genetically deleting Panx1 in male mouse models of AKI attenuates renal senescence and fibrosis. These findings, validated in human kidney tissue, identify ER-resident Panx1 as a critical driver of kidney disease progression and a potential therapeutic target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Kellum, J. A. et al. Acute kidney injury. Nat. Rev. Dis. Prim.7, 52 (2021). - PubMed
-
- Kellum, J. A., Ronco, C. & Bellomo, R. Conceptual advances and evolving terminology in acute kidney disease. Nat. Rev. Nephrol.17, 493–502 (2021). - PubMed
-
- Liu, B. C., Tang, T. T., Lv, L. L. & Lan, H. Y. Renal tubule injury: a driving force toward chronic kidney disease. Kidney Int.93, 568–579 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

