Bioengineered hybrid dual-targeting nanoparticles reprogram the tumour microenvironment for deep glioblastoma photodynamic therapy
- PMID: 40826003
- PMCID: PMC12361390
- DOI: 10.1038/s41467-025-63081-2
Bioengineered hybrid dual-targeting nanoparticles reprogram the tumour microenvironment for deep glioblastoma photodynamic therapy
Abstract
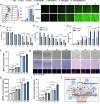
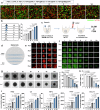
Glioblastoma (GBM) poses significant therapeutic challenges due to its hypoxic and immunosuppressive tumour microenvironment (TME), low immunogenicity and physical barriers. While combining photodynamic therapy (PDT) with immunotherapy holds promise, its efficacy is hampered by insufficient immune activation. In this study, we develop a multifunctional photodynamic-enhanced biomimetic intelligent nanoplatform (FBFO@HM@aOPN) responsive to the TME. The nanoplatform consists of a dual-enzyme nanozyme encapsulated in a prokaryotic-eukaryotic hybrid membrane, further modified with a pH-sensitive tumor-targeting antibody. After systemic administration, FBFO@HM@aOPN selectively accumulates in the GBM through vascular regulation and extracellular matrix (ECM) remodelling while generating oxygen to alleviate hypoxia. Crucially, the platform concurrently induces immunogenic death in tumour cells and reprograms protumoral macrophages to antitumor phenotypes. This dual action robustly activates both innate and adaptive immunity, significantly inhibiting GBM growth. Furthermore, when combined with anti-PD1 immunotherapy, the nanoplatform dramatically boosts the treatment effect and effectively prevents postsurgical tumour recurrence. Therefore, our work offers a multimodal platform for stimulating anti-tumour immunity, with potential applicability for GBM patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare competing interests.
Figures






References
-
- Meric-Bernstam, F., Larkin, J., Tabernero, J. & Bonini, C. Enhancing anti-tumour efficacy with immunotherapy combinations. Lancet397, 1010–1022 (2021). - PubMed
-
- Venkataramani, V. et al. Disconnecting multicellular networks in brain tumours. Nat. Rev. Cancer22, 481–491 (2022). - PubMed
-
- Wang, A. P. A review of glioblastoma and other primary brain malignancies. JAMA330, 188–189 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

