Characterization of a MERS-related betacoronavirus in Danish brown long-eared bats (Plecotus auritus)
- PMID: 40826422
- PMCID: PMC12362977
- DOI: 10.1186/s12985-025-02883-8
Characterization of a MERS-related betacoronavirus in Danish brown long-eared bats (Plecotus auritus)
Abstract
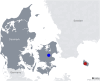
Background: Bats are recognized as natural reservoir hosts for numerous viruses and are believed to be the evolutionary origin of alpha- and beta-coronaviruses (CoVs), such as SARS-CoV, SARS-CoV-2, and possibly MERS-CoV. MERS-related beta-CoVs have been identified in bat species from Africa, America, Asia, and Europe. In this study, we describe the first detection and characterization of a MERS-related beta-CoV in Danish brown long-eared bats (Plecotus auritus).
Methods: Fecal samples collected through a national surveillance program were screened using pan-CoV RT-qPCRs. Positive samples underwent ORF1b sequencing, microarray analysis and Illumina MiSeq sequencing, followed by metagenomic assembly of full-length genomes. A global phylogenetic tree was used to determine placement within the Coronaviridae family and local maximum likelihood phylogenetic analysis clarified subgroup placement. The receptor-binding potential of the spike protein to human DPP4, ACE2, and bat ACE2 orthologs was assessed through phylogenetic analysis of the receptor-binding domain (RBD), alongside homology modeling and structural analysis.
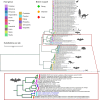
Results: Three samples tested positive for CoVs. One sample from a Soprano pipistrelle (Pipistrellus pygmaeus) was identified as alpha-CoV by ORF1b sequencing. The remaining two samples, obtained from a colony of Plecotus auritus, were identified as beta-CoVs, and separate microarray results indicated the presence of a MERS-related CoV. Full genomes were successfully assembled using a metagenomic approach. Phylogenetic analysis placed them within the merbecoviruses, forming a distinct clade with viruses detected in Vespertilionidae bats from Western Europe and East Asia. Analysis of the RBD placed them within the HKU25 clade. Structural modeling suggested hydrogen bonding patterns between the RBD and human/bat ACE2 orthologs or human DPP4, similar to known in vitro complexes, indicating potential receptor binding.
Conclusion: This is the first report of MERS-related beta-CoVs in bats from Denmark. Phylogenetic analyses reveal that these novel viruses belong to the HKU25 clade, a clade with known ACE2 receptor preference. Experimental validation is needed to confirm the receptor-binding potential, as additional interactions at the RBD-receptor interface may differ from previously described bat-merbecoviruses. Continued surveillance is crucial to identify potential intermediate hosts and assess interspecies transmission risk, with focus on the spike protein, receptor specificity, and binding affinity.
Keywords: Bats; Betacoronavirus; Denmark; Full genome sequencing; MERS-related; Merbecovirus; Molecular analysis; Phylogenetic analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical review and approval were waived for this study, since the only animal samples used were fecal samples collected without causing harm to the bats. The collection was conducted under dispensation MST-850-00116 from the Danish Environmental Protection Agency. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
In vitro and in vivo characterization of a bat merbecovirus with ACE2- and DPP4-independent cell entry.J Virol. 2025 Jul 22;99(7):e0072725. doi: 10.1128/jvi.00727-25. Epub 2025 Jun 17. J Virol. 2025. PMID: 40525828 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Detection and Prevalence of Coronaviruses in European Bats: A Systematic Review.Ecohealth. 2024 Dec;21(2-4):125-140. doi: 10.1007/s10393-024-01688-5. Epub 2024 Nov 23. Ecohealth. 2024. PMID: 39580592 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
References
-
- Burgin CJ, Colella JP, Kahn PL, Upham NS. How many species of mammals are there? J Mammal. 2018;99:1–14. 10.1093/jmammal/gyx147.
-
- Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chrétien D, Sanamxay D, et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022;604:330–6. 10.1038/s41586-022-04532-4. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

