Topology-aware pathway analysis of spatial transcriptomics
- PMID: 40827204
- PMCID: PMC12358110
- DOI: 10.7717/peerj.19729
Topology-aware pathway analysis of spatial transcriptomics
Abstract
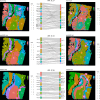
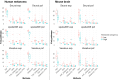
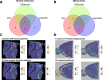
Spatial transcriptomics (ST) has transformed genomics by mapping gene expression onto intact tissue architecture, uncovering intricate cellular interactions that bulk and single-cell RNA sequencing often overlook. Traditional ST workflows typically involve clustering spots, performing differential expression analyses, and annotating results via gene-set methods such as overrepresentation analysis (ORA) or gene set enrichment analysis (GSEA). More recent spatially-aware techniques extend these approaches by incorporating tissue organization into gene-set scoring. However, because they operate primarily at the level of individual genes, they may overlook the connectivity and topology of biological pathways, limiting their capacity to trace the propagation of signaling events within tissue regions. In this study, we address that gap by translating gene expression into pathway-level activity using the Pathway Signal Flow (PSF) algorithm. PSF integrates expression data with curated interaction networks to compute numeric activity scores for each branch of a biological pathway, producing a functionally annotated feature space that captures downstream signaling effects as branch-specific activity values. We applied PSF to two public 10x Genomics Visium datasets (human melanoma and mouse brain) and compared clustering based on PSF-derived pathway activities from 40 curated Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways and gene expression with standard Seurat Louvain clustering and spatially aware methods (Vesalius, spatialGE). We observed good correspondence between PSF-based and expression-based clustering when spatially aware clustering methods were used. This suggests that branch-level pathway activities can themselves drive clustering and pinpoint spatially deregulated processes. To assess cluster-specific functional annotation, we compared PSF results to conventional ORA (based on marker genes) and GSDensity (based on cluster-specific gene sets). PSF identified a broader set of significant pathways with substantial overlap with both ORA and GSDensity, providing increased sensitivity due to its branch-level resolution. We further demonstrated that PSF-derived activity values can be used to detect spatially deregulated pathway branches, yielding results comparable to those obtained with spatially aware gene set analysis approaches such as GSDensity and spatialGE. The availability of pathway topology and branch-specific information also enabled the identification of potential intercellular communication via ligand-receptor interactions between deregulated pathways in adjacent tumor regions. To support interactive exploration of results, we developed the PSF Spatial Browser, an R Shiny application for visualizing pathway activities, gene expression patterns, and deregulated pathway networks.
Keywords: 10x Visium; Cancer omics; Cell–cell interaction; Functional analysis; Melanoma; Signaling pathways; Spatial transcriptomics; Topology-aware pathway analysis; Tumour microenvironment; Visualization.
©2025 Hakobyan et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

