Genome evolution of Acinetobacter baylyi ADP1 during laboratory domestication: acquired mutations impact competence and metabolism
- PMID: 40827875
- PMCID: PMC12442408
- DOI: 10.1128/aem.00936-25
Genome evolution of Acinetobacter baylyi ADP1 during laboratory domestication: acquired mutations impact competence and metabolism
Abstract
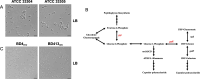
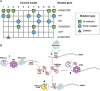
The bacterium Acinetobacter baylyi is a model organism known for its extreme natural competence and metabolic versatility. It is capable of taking up environmental DNA at a high rate across all growth phases. The type strain ADP1 was created by random mutagenesis of a precursor strain, BD4, to prevent it from forming cell chains in culture. ADP1 has since been distributed between research groups over several decades and acquired subsequent mutations during this time. In this study, we compare the genome sequences of A. baylyi BD4 and its modern descendants to identify and understand the effects of mutations acquired and engineered during its domestication. We demonstrate that the ADP1 variants in use today differ in their competence, growth on different carbon sources, and autoaggregation. In addition, we link the global carbon storage regulator CsrA and a transposon insertion that removes its C-terminal domain specifically to changes in both overall competence and an almost complete loss of competence during the stationary phase. Reconstructing the history of ADP1 and the diversity that has evolved in the variants currently in use improves our understanding of the desirable properties of this experimentally and industrially important bacterium and suggests ways that its reliability can be improved through further genome engineering.IMPORTANCEAcinetobacter baylyi ADP1 is a bacterial chassis of interest to microbiologists in academia and industry due to its extreme natural competence and wide metabolic range. Its ability to take up DNA from its environment makes it straightforward to efficiently edit its chromosome. We identify and characterize mutations that have been passed down to modern strains of ADP1 from the initial work in the 1960s, as well as subsequent mutations and genome edits separating strains in use by different research groups today. These mutations, including one in a global regulator (CsrA), have significant phenotypic consequences that have affected the reproducibility and consistency of experiments reported in the literature. We link a mutation in this global regulator to unexpected changes in natural competence. We also show that domesticated A. baylyi strains have impaired growth on a variety of carbon sources.
Keywords: Acinetobacter baylyi; domestication; laboratory evolution; natural competence; post-transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

