Detergent-free isolation and characterization of amyloid precursor protein C99 in E. coli native lipid-nanodiscs using non-ionic polymer
- PMID: 40828514
- PMCID: PMC12363408
- DOI: 10.1002/pro.70276
Detergent-free isolation and characterization of amyloid precursor protein C99 in E. coli native lipid-nanodiscs using non-ionic polymer
Abstract
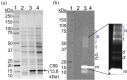
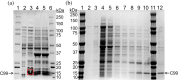
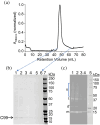
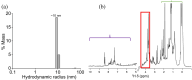
Alzheimer's disease (AD), a progressive neurodegenerative disorder, is characterized by cognitive decline resulting from neuronal cell death. A key contributor to AD pathology is C99, a membrane-bound β-secretase-cleaved fragment of amyloid precursor protein (APP). C99 plays a central role in generating amyloid-beta (Aβ) isomers, which are directly implicated in disease progression. Understanding its structure and lipid interactions is essential for elucidating its mechanistic role in AD and guiding therapeutic development. C99 has been studied in membrane mimetics such as micelles, bicelles, and reconstituted nanodiscs. Although reconstituted nanodiscs provide a native-like lipid-bilayer environment, the use of detergents prior to reconstitution has been reported to disrupt native folding and lipid-protein interactions. In this study, we successfully isolated and purified C99 along with its associated lipids directly from E. coli cell membranes using a non-ionic pentyl-inulin polymer, avoiding the need for detergents. The purified C99-containing pentyl-inulin nanodiscs were characterized using SDS-PAGE, Western blotting, dynamic light scattering (DLS), 1H NMR spectroscopy, matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry, and liquid chromatography-mass spectrometry (LC-MS). Notably, we observed SDS-stable oligomers of C99. DLS and 1H NMR confirmed the presence of large particles composed of pentyl-inulin and E. coli lipids. MALDI-TOF and LC-MS verified the molecular mass and amino acid sequence of C99, respectively. We propose that this detergent-free method for the direct isolation of C99 and native lipids using non-ionic pentyl-inulin may serve as a valuable tool for investigating the C99-secretase complex and for developing compounds aimed at inhibiting the production of amyloid-beta isomers.
Keywords: Alzheimer's disease; C99 oligomer; amyloid precursor protein (APP‐C99); detergent‐free purification; nanodisc; non‐ionic polymer.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures
Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
A C. elegans model of familial Alzheimer's disease shows age-dependent synaptic degeneration independent of amyloid β-peptide.bioRxiv [Preprint]. 2025 Jul 20:2025.07.16.665161. doi: 10.1101/2025.07.16.665161. bioRxiv. 2025. PMID: 40791515 Free PMC article. Preprint.
-
Clear Native Gel Electrophoresis for the Purification of Fluorescently Labeled Membrane Proteins in Native Nanodiscs.Anal Chem. 2025 Aug 12;97(31):16796-16804. doi: 10.1021/acs.analchem.5c01702. Epub 2025 Aug 1. Anal Chem. 2025. PMID: 40748368 Free PMC article.
-
18F PET with florbetapir for the early diagnosis of Alzheimer's disease dementia and other dementias in people with mild cognitive impairment (MCI).Cochrane Database Syst Rev. 2017 Nov 22;11(11):CD012216. doi: 10.1002/14651858.CD012216.pub2. Cochrane Database Syst Rev. 2017. PMID: 29164603 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Bao H, Dalal K, Wang V, Rouiller I, Duong F. The maltose ABC transporter: action of membrane lipids on the transporter stability, coupling and ATPase activity. Biochimica et Biophysica Acta. 2013;1828:1723–1730. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

