Integrative proteogenomic characterization of Wilms tumor
- PMID: 40830093
- PMCID: PMC12365320
- DOI: 10.1038/s41467-025-62234-7
Integrative proteogenomic characterization of Wilms tumor
Abstract
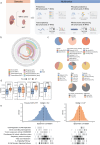
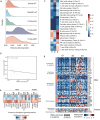
Wilms tumor (WT), the most common pediatric renal malignancy, exhibits a relatively low mutational burden compared to adult cancers, which hinders the development of targeted therapies. To elucidate the molecular landscape of WT, we perform integrative proteomic, phosphoproteomic, transcriptomic, and whole-exome sequencing analyses of WT and normal kidney tissue adjacent to tumor. Our multi-omics approach uncovers prognostic genetic alterations, distinct molecular subgroups, immune microenvironment features, and potential biomarkers and therapeutic targets. Proteome- and transcriptome-based stratification identifies three molecular subgroups with unique signatures, correlating with different histopathological subtypes and putative cellular origins at different stages of embryonic kidney development. Notably, we identify EHMT2 as a promising prognostic biomarker and therapeutic target associated with epigenetic regulation and Wnt/β-catenin pathway. In this work, we provide a comprehensive molecular characterization of WT, offering valuable insights into its pathogenesis and a foundational resource for future therapeutic development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All authors declare no competing interests.
Figures





References
-
- Breslow, N., Olshan, A., Beckwith, J. B. & Green, D. M. Epidemiology of Wilms tumor. Med. Pediatr. Oncol.21, 172–181 (1993). - PubMed
-
- Ni, X. et al. Socioeconomic inequalities in cancer incidence and access to health services among children and adolescents in China: a cross-sectional study. Lancet400, 1020–1032 (2022). - PubMed
-
- Wilms Tumor and Other Childhood Kidney Tumors Treatment (PDQ(R)): health Professional Version. In: PDQ Cancer Information Summaries) (PDQ, 2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

