Extremely acidophilic filamentous fungi are more prevalent in diverse ecosystems than previously documented
- PMID: 40830373
- PMCID: PMC12365195
- DOI: 10.1038/s41598-025-06321-1
Extremely acidophilic filamentous fungi are more prevalent in diverse ecosystems than previously documented
Abstract
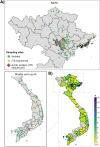
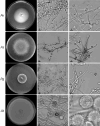
The study of extremophiles can lead to the discovery of highly tolerant enzymes of value to biotechnology, and extreme environments are typically sampled to facilitate their discovery. Here, we show that extreme acidotolerant filamentous fungi, able to grow at pH < 1, can be found when sampling highly diverse environments, from industrial sites with low pH to typical non-acidic plant and soil samples. Over 100 fungal strains were isolated from over 2,000 samples taken from across Vietnam, and many of the strains were able to grow over wide pH ranges, displaying either acidotolerance or clear acidophilicity. ITS sequencing revealed that 63 isolates represent 12 previously undescribed species, with the majority from the Talaromyces or Penicillium genera. We furthermore report the rediscovery of the previously lost, historically significant acidophile Acontium velatum. Screening of selected fungal secretomes for polysaccharide-cleaving activity revealed that many show broad tolerance to harsh conditions (pH, temperature, organic solvents). Our work greatly expands on the diversity of identified extreme acidotolerant and acidophilic filamentous fungi, which can serve as sources of industrially relevant enzymes. For most species identified, acid tolerance or acidophilicity has not previously been reported, and our results showcase that acidophilicity is more widespread than previously appreciated.
Keywords: Acontium; Penicillium; Talaromyces; Acidophile; Acidotolerant; Ascomycota; Biodiversity; CAZymes; Extremophile; New fungal taxa.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Adherence to national and international regulations: All work described in this manuscript has been performed under a memorandum of understanding between FIRI and Chalmers University of Technology. Competing interests: The authors declare no competing interests.
Figures



References
-
- Seckbach, J. Life on the edge and astrobiology: who is who in the polyextremophiles world? In Polyextremophiles: Life Under Multiple Forms of Stress (eds Seckbach, J. et al.) 63–79 (Springer, 2013).
-
- Johnson, D. B. Physiology and ecology of acidophilic microorganisms. Physiol. Biochem. Extremophiles. 255–270 (2014).
-
- Baker-Austin, C. & Dopson, M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol.15(4), 165–171 (2007). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

