Machine learning based screening of biomarkers associated with cell death and immunosuppression of multiple life stages sepsis populations
- PMID: 40830558
- PMCID: PMC12365015
- DOI: 10.1038/s41598-025-14600-0
Machine learning based screening of biomarkers associated with cell death and immunosuppression of multiple life stages sepsis populations
Abstract
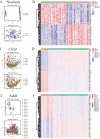
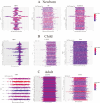
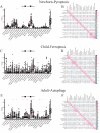
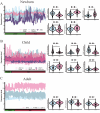
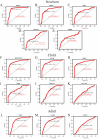
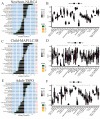
Sepsis is a condition resulting from the uncontrolled immune response to infection, leading to widespread inflammatory damage and potentially fatal organ dysfunction. Currently, there is a lack of specific prevention and treatment strategies for sepsis across different age groups. Programmed Cell Death (PCD) can regulate the enrichment of effector immune cells or regulatory immune cells, providing a new perspective for immunotherapy. Within the framework of computational biology and machine learning strategies, and against the backdrop of global multicenter sepsis cohort data, this study aims to deeply mine and screen specific biomarkers related to the immune microenvironment and programmed cell death in populations across different life stages (neonates, children, and adults). This will provide foundational data for precision treatment and drug development in artificial intelligence-assisted sepsis diagnosis and treatment management. Gene expression data from sepsis patients across global multicenter populations, including China, Europe, and the United States, were obtained from the Gene Expression Omnibus (GEO) database, and differentially expressed genes (DEGs) were identified. A literature review was conducted to obtain 18 PCD-related genes, which were intersected with DEGs to identify DEGs associated with specific types of PCD. Nine machine learning algorithms (Logistic Regression LR, Decision Tree DT, Gradient Boosting Machine GBM, K-Nearest Neighbors KNN, LASSO, Principal Component Analysis PCA, Random Forest RF, Support Vector Machine SVM, and XGBoost) were applied to training and testing datasets with 10-fold cross-validation to select three optimized algorithm models. The SHAP algorithm was further used to quantify the contribution of each gene based on cell death features to the prediction of sepsis. Key PCD patterns were identified based on model evaluation metrics (Accuracy, Precision, Recall, F1 score, and Receiver Operating Characteristic Curve ROC), and their associated DEGs were obtained through intersection, followed by immune-related analysis of DEGs. The study included a total of 1507 sepsis cases and 484 controls globally, with 90 neonatal cases and 95 controls, 527 children cases and 101 controls, and 890 adult cases and 288 controls. The best model for predicting sepsis across different populations was GBM.The key PCD patterns selected by machine learning for different age groups were Pyroptosis (neonates), Ferroptosis (children), and Autophagy (adults). (1) In neonatal sepsis, the models constructed by GBM, XGBoost, and RF algorithms performed the best, and identified 5 key DEGs associated with Pyroptosis (CHMP7, NLRC4, AIM2, GZMB, PRKACA), with NLRC4 showing the best predictive ability (AUC = 0.902, P < 0.05), significantly positively correlated with neutrophils and negatively correlated with CD8 + T cells. (2) In the children sepsis population, models constructed using the Gradient Boosting Machine (GBM), Support Vector Machine (SVM), and Least Absolute Shrinkage and Selection Operator (LASSO) algorithms demonstrated the best performance. Six key DEGs associated with Ferroptosis were identified (AKR1C3, GCLM, PEBP1, CARS, MAP1LC3B, SCL11A2), among which MAP1LC3B, playing a role in mitochondrial reactive oxygen species energy metabolism, showed the strongest predictive ability (AUC = 0.883, P < 0.05). It was significantly positively correlated with M0-type macrophages and significantly negatively correlated with activated CD4 + memory T cells. (3) In the adult sepsis population, models constructed using GBM, SVM, and LASSO algorithms showed the best performance. Three key DEGs associated with Autophagy were identified (TSPO, HTRA2, USP10), with TSPO, which mediates oxidative stress regulation, iron homeostasis, and cholesterol transport, showing the strongest predictive ability (AUC = 0.825, P < 0.05). It was significantly positively correlated with M1-type macrophages and significantly negatively correlated with CD8 + T cells. This study, through the integrated application of computational biology and machine learning algorithms, discovered biomarkers of PCD patterns that affect cytokine storm-mediated inflammation and immunosuppressive effects in sepsis populations across different age groups (neonates, children, and adults). These findings have specific clinical application and drug development value, providing a scientific basis for the global application of artificial intelligence-assisted sepsis diagnosis and treatment management.
Keywords: Biomarkers; Cell death; Immunosuppression; Machine learning; Sepsis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Supervised Machine Learning Models for Predicting Sepsis-Associated Liver Injury in Patients With Sepsis: Development and Validation Study Based on a Multicenter Cohort Study.J Med Internet Res. 2025 May 26;27:e66733. doi: 10.2196/66733. J Med Internet Res. 2025. PMID: 40418571 Free PMC article.
-
Molecular subtypes of lung adenocarcinoma patients for prognosis and therapeutic response prediction with machine learning on 13 programmed cell death patterns.J Cancer Res Clin Oncol. 2023 Oct;149(13):11351-11368. doi: 10.1007/s00432-023-05000-w. Epub 2023 Jun 28. J Cancer Res Clin Oncol. 2023. PMID: 37378675 Free PMC article.
-
Construction and validation of HBV-ACLF bacterial infection diagnosis model based on machine learning.BMC Infect Dis. 2025 Jul 1;25(1):847. doi: 10.1186/s12879-025-11199-5. BMC Infect Dis. 2025. PMID: 40596896 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Disulfidptosis: A new type of cell death.Apoptosis. 2024 Oct;29(9-10):1309-1329. doi: 10.1007/s10495-024-01989-8. Epub 2024 Jun 17. Apoptosis. 2024. PMID: 38886311 Free PMC article. Review.
References
-
- Neto, A. S., Dessap, A. M. & Papazian, L. Focus on ARDS. Intensive Care Med.43(10), 1495–1497 (2017). - PubMed
-
- Tamayo, E. et al. Pro- and anti-inflammatory responses are regulated simultaneously from the first moments of septic shock. Eur. Cytokine Netw.22(2), 82–87 (2011). - PubMed
-
- Gaieski, D. F. et al. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit. Care Med.41(5), 1167–1174 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

