Multiplexed bacteriocin synthesis to combat and prevent antimicrobial resistance
- PMID: 40830650
- PMCID: PMC12365309
- DOI: 10.1038/s42003-025-08639-y
Multiplexed bacteriocin synthesis to combat and prevent antimicrobial resistance
Abstract
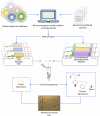
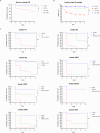
Bacteriocins are underexplored yet promising candidates to combat antimicrobial resistance (AMR) and enable targeted therapy due to their natural origin, abundance and narrow spectrum of activity. In this study, we used a collection of engineered DNA devices and cell-free gene expression (CFE) to rapidly produce combinations (cocktails) of bacteriocins comprising both linear and circular proteins. Other cocktails were designed to target a specific bacterial species by leveraging insights into bacteriocin pathways for cell envelope penetration. These tailored combinations eradicated bacteria effectively while preventing resistance development. The synthesis of bacteriocins was optimized by using continuous exchange CFE, reengineering DNA parts, and adjusting conditions for disulfide bond formation. Also, we illustrate the efficacy of these bacteriocin mixtures against various multidrug-resistant human pathogens and highlight their potential through in vivo testing in the animal model Galleria mellonella. Our bacteriocin cocktail expression and test platform underscores the potential of bacteriocins for innovative treatments against multidrug-resistant infections.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following competing interests: Juan Borrero (JB), Pascal Hols (PH) and Philippe Gabant (PG) declare that they are listed as inventors in the following patents and patent applications related to bacteriocin production and uses: J.B. and P.G., “Bacteriocin polypeptides, nucleic acids encoding same, and methods of use thereof”, Application PCT/US2023/067567; P.G., “Controlled growth of microorganisms”, US Patents 9,333,227/10,188,114/11,427,800/12,297,422 + CN ZL 201480057387.2/ZL 201910882176.7 + EP3035802B1 + BR112016003533-0/1220210154171 + IN389267; P.H.: “Peptides for inducing bacteriocin synthesis and methods to identify and/or select and/or optimize the same”, US Patent 12,234,299. P.H. is a member of the Scientific Advisory Board of the Syngulon company. All other authors declare no competing interests.
Figures





References
-
- Cotter, P. D., Ross, R. P. & Hill, C. Bacteriocins—a viable alternative to antibiotics?. Nat. Rev. Microbiol.11, 95–105 (2013). - PubMed
-
- Heilbronner, S., Krismer, B., Brötz-Oesterhelt, H. & Peschel, A. The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol.19, 726–739 (2021). - PubMed
-
- Guinane, C. M. et al. The bacteriocin bactofencin A subtly modulates gut microbial populations. Anaerobe40, 41–49 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

