RBM15 enhances paclitaxel resistance in triple-negative breast cancer by targeting m6A methylation of TNFSF9 and inducing polarization of tumor-associated macrophages to M2 phenotype
- PMID: 40830812
- PMCID: PMC12362948
- DOI: 10.1186/s41065-025-00534-0
RBM15 enhances paclitaxel resistance in triple-negative breast cancer by targeting m6A methylation of TNFSF9 and inducing polarization of tumor-associated macrophages to M2 phenotype
Abstract
Background: Triple-negative breast cancer (TNBC) is one of the breast cancer subtypes with a poor prognosis, and the current main treatment modalities include surgical resection and adjuvant chemotherapy. However, the development of drug resistance in tumor cells to chemotherapeutic agents poses great challenges to anticancer treatment.
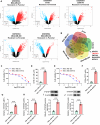
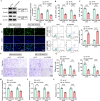
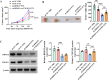
Methods: Bioinformatics analysis was used to screen the up-regulated genes in paclitaxel (PTX)-resistant TNBC cells. Cell viability was measured by a CCK-8 kit. TNFSF9 (Tumor necrosis factor receptor superfamily member 9) protein level was detected by Western blot (WB) assay. PTX-resistant TNBC cell lines (MDA-MB-231/PTX, MDA-MB-468/PTX) were constructed and their drug resistance was shown by IC50. The EdU, flow cytometry, Transwell, and other commercial kits were applied to detect the proliferation, apoptosis, migration, invasion, macrophage M2 polarization, and glycolysis of PTX-resistant TNBC cells. RBM15 (RNA binding motif protein 15) levels were measured by RT-qPCR and WB assays. The RIP, MeRIP, and actinomycin D assays were used to analyze the interaction between TNFSF9 and RBM15. The effect of RBM15/TNFSF9 on PTX sensitivity in vivo was verified by xenograft tumor experiments.
Results: TNFSF9 was highly expressed in PTX-resistant TNBC cells. Silencing of TNFSF9 enhanced the sensitivity to PTX. Silencing TNFSF9 induced polarization of macrophages from M2 to M1 phenotype and the release of IL-1β and TNF-α, but decreased the levels of IL-10 and TGF-β. RBM15 targeted the N6-adenylate methylation (m6A) modification of TNFSF9, and overexpression of TNFSF9 could reverse the tumor-suppressing effect of silencing RBM15 on PTX-resistant TNBC cells in vitro and transplanted tumors in vivo. Samples from PTX-sensitive and PTX-resistant TNBC patients proved that RBM15 regulated TNFSF9's high expression in PTX-resistant TNBC tissues.
Conclusion: This study demonstrated that RBM15 enhanced PTX resistance in TNBC by promoting m6A methylation in TNFSF9 and inducing M2 polarization of tumor-associated macrophages.
Keywords: M2 polarization; PTX-resistant; RBM15; TNFSF9; Triple-negative breast cancer.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The present study was approved by the ethical review committee of Xianyang Hospital of Yan'an University with approval No.20240128. Written informed consent was obtained from all enrolled patients. Patients agree to participate in this work Competing interests: The authors declare no competing interests. Disclosure of interest: The authors declare that they have no financial or non-financial conflicts of interest.
Figures





Similar articles
-
The IL-8/NF-κB feedback loop confers a paclitaxel-sensitive/doxorubicin-resistant phenotype in triple-negative breast cancer.Free Radic Biol Med. 2025 Oct;238:316-328. doi: 10.1016/j.freeradbiomed.2025.06.049. Epub 2025 Jun 27. Free Radic Biol Med. 2025. PMID: 40582426
-
Zhuidu Formula suppresses the migratory and invasive properties of triple-negative breast cancer cells via dual signaling pathways of RhoA/ROCK and CDC42/MRCK.J Ethnopharmacol. 2023 Oct 28;315:116644. doi: 10.1016/j.jep.2023.116644. Epub 2023 May 16. J Ethnopharmacol. 2023. PMID: 37196814
-
SNHG14 promotes triple-negative breast cancer cell proliferation, invasion, and chemoresistance by regulating the ERK/MAPK signaling pathway.IUBMB Life. 2024 Dec;76(12):1295-1308. doi: 10.1002/iub.2910. Epub 2024 Sep 12. IUBMB Life. 2024. PMID: 39266460
-
The role of long non-coding RNAs in developing paclitaxel-resistant triple negative breast cancer: a systematic review.Cancer Treat Res Commun. 2025;43:100936. doi: 10.1016/j.ctarc.2025.100936. Epub 2025 Apr 30. Cancer Treat Res Commun. 2025. PMID: 40344739
-
An update on cancer stem cell survival pathways involved in chemoresistance in triple-negative breast cancer.Future Oncol. 2025 Mar;21(6):715-735. doi: 10.1080/14796694.2025.2461443. Epub 2025 Feb 12. Future Oncol. 2025. PMID: 39936282 Review.
References
-
- Abdullah H, Neelaveni T, Mohammed A, et al. <i > In-silico binding, stability, pharmacokinetics, and toxicity studies on natural (-)-ambrox metabolites as binding ligands to luminal B and Triple-negative/basal-like proteins for breast cancer therapy</i >. Lett Drug Des Discovery. 2024;21:1569–81.
-
- Jiangqin Z, Chunhui T, Xun F, Mei Q, Yuewen J. Deciphering the Multi-target Pharmacological mechanism of Sojae semen nigrum acting on breast cancer through integrating network Pharmacology and molecular Docking approaches. Lett Drug Des Discovery. 2024;21:4565–86.
-
- Ayse Gülbin K, Ihsan K, Ahmet S, Seniz D, Abdullah Tuncay D. Impact of radiation therapy on serum humanin and MOTS-c levels in patients with lung or breast cancer. Curr Radiopharmaceuticals. 2024;17:229–37. - PubMed
-
- Yasaman N, Zahra H, Vahid R, Sahar K. Deregulated MicroRNAs involved in P53 signaling pathway in breast cancer with focus on Triple-negative breast cancer. Curr Mol Pharmacol. 2024;17:1–20. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

