Multimodal analysis of cell-free DNA enhances differentiation of early-stage breast cancer from benign lesions and healthy individuals
- PMID: 40830871
- PMCID: PMC12366216
- DOI: 10.1186/s12915-025-02371-z
Multimodal analysis of cell-free DNA enhances differentiation of early-stage breast cancer from benign lesions and healthy individuals
Abstract
Background: Breast cancer (BC) remains the second leading cause of cancer-related mortality among women worldwide. Liquid biopsy based on circulating tumor DNA (ctDNA) offers a promising noninvasive approach for early detection; however, differentiating malignant tumors from benign abnormalities remains a significant challenge.
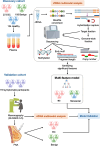
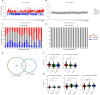
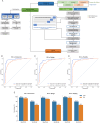
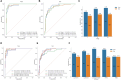
Results: Here, we developed a multimodal approach to analyze cfDNA methylation and fragmentomic patterns in 273 BC patients, 108 individuals with benign breast conditions, and 134 healthy controls. Genome-wide analyses revealed distinct cfDNA copy number alterations and cytosine-enriched cleavage sites in BC patients. Targeted sequencing further revealed unique methylation patterns, including hypermethylation in GPR126, KLF3, and TLR10 and hypomethylation in TOP1 and MAFB. Our machine-learning model achieved an AUC of 0.90, with 93.6% specificity and 62.1-66.3% sensitivity for stage I-II cancers. In symptomatic populations, sensitivities were 50.0%, 68.2%, and 64.7% for BI-RADS categories 3, 4, and 5, respectively, with 96.1% specificity.
Conclusions: These findings underscore the potential of cfDNA biomarkers to enhance BC detection and reduce the rate of unnecessary biopsies.
Keywords: Benign abnormalities; Breast cancer; CfDNA; Methylation and fragmentomic.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The research protocol was approved by the Ethics Committees of all participating institutions, with ethics review number 294/BVUB-HĐĐĐ for the discovery cohort and 460/HĐĐĐ-ĐHYD for the validation cohort. Informed written consent was obtained from each participant in accordance with the Declaration of Helsinki. Consent for publication: Not applicable. Competing interests: The authors, including LST, DSN, HG, MDP, and HNN, hold equity in Gene Solutions. We confirm that this does not impact on our compliance with the journal's policies regarding data and material sharing.
Figures


References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–63. - PubMed
-
- Merino Bonilla JA, Torres Tabanera M, Ros Mendoza LH. Breast cancer in the 21st century: from early detection to new therapies. Radiologia. 2017;59(5):368–79. - PubMed
-
- Gilbert FJ, Tucker L, Young KC. Digital breast tomosynthesis (DBT): a review of the evidence for use as a screening tool. Clin Radiol. 2016;71(2):141–50. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

