A message passing framework for precise cell state identification with scClassify2
- PMID: 40830895
- PMCID: PMC12362893
- DOI: 10.1186/s13059-025-03722-3
A message passing framework for precise cell state identification with scClassify2
Abstract
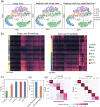
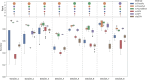
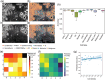
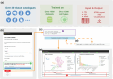
Cell annotation is crucial for downstream exploration. Although many approaches, spanning from classic statistics to large language models, have been developed, most of their focus is on distinct cell types and overlook sequential cell populations. Here, we propose an annotation method, scClassify2, to specifically focus on adjacent cell state identification. By incorporating prior biological knowledge through a novel dual-layer architecture and ordinal regression, scClassify2 achieves competitive performance compared to other state-of-the-art methods. Besides single-cell RNA-sequencing data, scClassify2 is generalizable from different platforms including subcellular spatial transcriptomics data. We also develop a web server for academic uses.
Keywords: Cell state identification; Dual layer architecture; MPNN; Ordinal regression; ScRNA-seq.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
ScInfeR: an efficient method for annotating cell types and sub-types in single-cell RNA-seq, ATAC-seq, and spatial omics.Brief Bioinform. 2025 May 1;26(3):bbaf253. doi: 10.1093/bib/bbaf253. Brief Bioinform. 2025. PMID: 40471991 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Integration tools for scRNA-seq data and spatial transcriptomics sequencing data.Brief Funct Genomics. 2024 Jul 19;23(4):295-302. doi: 10.1093/bfgp/elae002. Brief Funct Genomics. 2024. PMID: 38267084 Review.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

