Inhaling Eugenol Inhibits NAFLD by Activating the Hepatic Ectopic Olfactory Receptor Olfr544 and Modulating the Gut Microbiota
- PMID: 40831283
- PMCID: PMC12622515
- DOI: 10.1002/advs.202510321
Inhaling Eugenol Inhibits NAFLD by Activating the Hepatic Ectopic Olfactory Receptor Olfr544 and Modulating the Gut Microbiota
Abstract
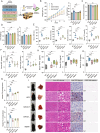
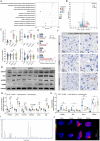
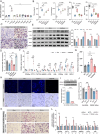
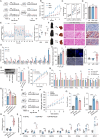
Non-alcoholic fatty liver disease (NAFLD) is a major public health threat with currently limited therapeutic options. Inhalation therapy shows promise for treating metabolic disorders due to rapid absorption and high patient adherence, though relevant medications remain scarce. Eugenol (EUG), the primary component of Syzygium aromaticum volatile oil, emerges as a promising NAFLD inhibitor from lipid-lowering aromatic Chinese medicine screening. EUG elicited significant anti-steatotic effects in both cultured hepatocytes and comprehensive high-fat diet-induced NAFLD animal models. Mechanistically, EUG targeted the activation of the hepatic ectopic olfactory receptor Olfr544 and up-regulated its downstream cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA)/cAMP response element binding protein signaling pathway, which further promoted fat lipolysis and oxidation. This effect is prevented by Olfr544 knockdown models both in vitro and in vivo, with supporting bioinformatics analysis. Moreover, EUG reversed gut microbiota dysbiosis and enriched two probiotic strains L. ruteri XR23 and L. johnsonii XR25, and oral gavage potently mitigated NAFLD in mice, with the key metabolites 3-indolepropionic acid (IPA) and 5-hydroxyindole-3-acetic acid (5-HIAA) inhibiting lipid synthesis. Lower levels of 5-HIAA and IPA are observed in patients with NAFLD. These results highlight the considerable potential of EUG as an agonist of Olfr544 for treating NAFLD by inhalation.
Keywords: NAFLD; eugenol; gut microbiota; olfactory receptor; volatile oils.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Wong V. W., Ekstedt M., Wong G. L., Hagström H., J. Hepatol. 2023, 79, 842. - PubMed
-
- Younossi Z. M., J. Hepatol. 2019, 70, 531. - PubMed
-
- Targher G., Tilg H., Byrne C. D., Lancet Gastroenterol. Hepatol. 2021, 6, 578. - PubMed
-
- Tilg H., Byrne C. D., Targher G., Lancet Gastroenterol. Hepatol. 2023, 8, 943. - PubMed
MeSH terms
Substances
Grants and funding
- 2022-6/National Qi-Huang scholar
- 82304831/National Natural Science Foundation of China
- 2023TQ0109 GZB20230196/China Postdoctoral Science Foundation
- 232301420019/Key Science and Technology Program of Henan Province
- 242300421090/Henan Science Fund for Excellent Young Scholars
- 232301420077/Associates Fund of Henan Province Science and Technology Research and Development Program
- 2025HYTP092/Young Talent Support Program of Henan Association for Science and Technology
- STG-ZYX02-202117 HSRP- DFCTCM-2023-3-13/Henan Province Traditional Chinese Medicine Double First-Class Scientific Research Project
LinkOut - more resources
Full Text Sources
Medical
