Multi-ancestral genome-wide association study of clinically defined nicotine dependence reveals strong genetic correlations with other substance use disorders and health-related traits
- PMID: 40831304
- PMCID: PMC12360691
- DOI: 10.1017/S0033291725100883
Multi-ancestral genome-wide association study of clinically defined nicotine dependence reveals strong genetic correlations with other substance use disorders and health-related traits
Abstract
Background: Genetic research on nicotine dependence has utilized multiple assessments that are in weak agreement.
Methods: We conducted a genome-wide association study (GWAS) of nicotine dependence defined using the Diagnostic and Statistical Manual of Mental Disorders (DSM-NicDep) in 61,861 individuals (47,884 of European ancestry [EUR], 10,231 of African ancestry, and 3,746 of East Asian ancestry) and compared the results to other nicotine-related phenotypes.
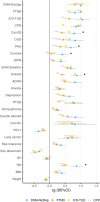
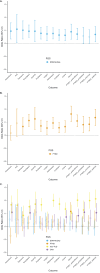
Results: We replicated the well-known association at the CHRNA5 locus (lead single-nucleotide polymorphism [SNP]: rs147144681, p = 1.27E-11 in EUR; lead SNP = rs2036527, p = 6.49e-13 in cross-ancestry analysis). DSM-NicDep showed strong positive genetic correlations with cannabis use disorder, opioid use disorder, problematic alcohol use, lung cancer, material deprivation, and several psychiatric disorders, and negative correlations with respiratory function and educational attainment. A polygenic score of DSM-NicDep predicted DSM-5 tobacco use disorder criterion count and all 11 individual diagnostic criteria in the independent National Epidemiologic Survey on Alcohol and Related Conditions-III sample. In genomic structural equation models, DSM-NicDep loaded more strongly on a previously identified factor of general addiction liability than a "problematic tobacco use" factor (a combination of cigarettes per day and nicotine dependence defined by the Fagerström Test for Nicotine Dependence). Finally, DSM-NicDep showed a strong genetic correlation with a GWAS of tobacco use disorder as defined in electronic health records (EHRs).
Conclusions: Our results suggest that combining the wide availability of diagnostic EHR data with nuanced criterion-level analyses of DSM tobacco use disorder may produce new insights into the genetics of this disorder.
Keywords: addiction; genome-wide association study; nicotine dependence; polygenic risk; psychiatric disorders.
Conflict of interest statement
The authors declare none.
Figures

Update of
-
Multi-ancestral genome-wide association study of clinically defined nicotine dependence reveals strong genetic correlations with other substance use disorders and health-related traits.medRxiv [Preprint]. 2025 Feb 3:2025.01.29.25320962. doi: 10.1101/2025.01.29.25320962. medRxiv. 2025. Update in: Psychol Med. 2025 Aug 20;55:e234. doi: 10.1017/S0033291725100883. PMID: 39974067 Free PMC article. Updated. Preprint.
Similar articles
-
Multi-ancestral genome-wide association study of clinically defined nicotine dependence reveals strong genetic correlations with other substance use disorders and health-related traits.medRxiv [Preprint]. 2025 Feb 3:2025.01.29.25320962. doi: 10.1101/2025.01.29.25320962. medRxiv. 2025. Update in: Psychol Med. 2025 Aug 20;55:e234. doi: 10.1017/S0033291725100883. PMID: 39974067 Free PMC article. Updated. Preprint.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Alcohol Use Disorder Polygenic Score Compared With Family History and ADH1B.JAMA Netw Open. 2024 Dec 2;7(12):e2452705. doi: 10.1001/jamanetworkopen.2024.52705. JAMA Netw Open. 2024. PMID: 39786404 Free PMC article.
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article.
-
Electronic cigarettes for smoking cessation.Cochrane Database Syst Rev. 2021 Sep 14;9(9):CD010216. doi: 10.1002/14651858.CD010216.pub6. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 Nov 17;11:CD010216. doi: 10.1002/14651858.CD010216.pub7. PMID: 34519354 Free PMC article. Updated.
References
MeSH terms
Substances
Grants and funding
- RC2 DA028909/DA/NIDA NIH HHS/United States
- K24 DA032555/DA/NIDA NIH HHS/United States
- N01 HG065403/HG/NHGRI NIH HHS/United States
- R01 MH077874/MH/NIMH NIH HHS/United States
- R01 AA009203/AA/NIAAA NIH HHS/United States
- R01 AA013321/AA/NIAAA NIH HHS/United States
- R01 DA018432/DA/NIDA NIH HHS/United States
- U01 HG004422/HG/NHGRI NIH HHS/United States
- R01 MH048085/MH/NIMH NIH HHS/United States
- R01 MH063671/MH/NIMH NIH HHS/United States
- R01 AA007535/AA/NIAAA NIH HHS/United States
- K23 MH080230/MH/NIMH NIH HHS/United States
- R01 DA035804/DA/NIDA NIH HHS/United States
- U01 HG004438/HG/NHGRI NIH HHS/United States
- R01 AA014041/AA/NIAAA NIH HHS/United States
- K05 AA017688/AA/NIAAA NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- R01 DA012690/DA/NIDA NIH HHS/United States
- R01 DA012849/DA/NIDA NIH HHS/United States
- P50 AA011998/AA/NIAAA NIH HHS/United States
- R01 MH063970/MH/NIMH NIH HHS/United States
- R01 AA007728/AA/NIAAA NIH HHS/United States
- R01 DA024413/DA/NIDA NIH HHS/United States
- R01 DA021905/DA/NIDA NIH HHS/United States
- R01 DA021913/DA/NIDA NIH HHS/United States
- R01 AA017444/AA/NIAAA NIH HHS/United States
- P01 CA089392/CA/NCI NIH HHS/United States
- U01 DA024413/DA/NIDA NIH HHS/United States
- R56 DA012854/DA/NIDA NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- K01 DA051759/DA/NIDA NIH HHS/United States
- R01 DA019963/DA/NIDA NIH HHS/United States
- R01 DA013423/DA/NIDA NIH HHS/United States
- K02 AA018755/AA/NIAAA NIH HHS/United States
- R01 DA012845/DA/NIDA NIH HHS/United States
- R01 AA011330/AA/NIAAA NIH HHS/United States
- R01 DA012854/DA/NIDA NIH HHS/United States
- K05 AA000145/AA/NIAAA NIH HHS/United States
- R00 DA023549/DA/NIDA NIH HHS/United States
- K08 DA019951/DA/NIDA NIH HHS/United States
- R01 AA017535/AA/NIAAA NIH HHS/United States
- K01 MH093731/MH/NIMH NIH HHS/United States
- R01 AA012502/AA/NIAAA NIH HHS/United States
- R01 DA017305/DA/NIDA NIH HHS/United States
- K01DA051759/DA/NIDA NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 DA011301/DA/NIDA NIH HHS/United States
- R01 AA015416/AA/NIAAA NIH HHS/United States
- U10 AA008401/AA/NIAAA NIH HHS/United States
- R37 AA007728/AA/NIAAA NIH HHS/United States
- R01 AA013320/AA/NIAAA NIH HHS/United States
- P60 DA011015/DA/NIDA NIH HHS/United States
- R37 AA012502/AA/NIAAA NIH HHS/United States

