This is a preprint.
Accelerating Biomolecular Modeling with AtomWorks and RF3
- PMID: 40832246
- PMCID: PMC12363939
- DOI: 10.1101/2025.08.14.670328
Accelerating Biomolecular Modeling with AtomWorks and RF3
Abstract
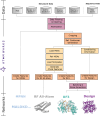
Deep learning methods trained on protein structure databases have revolutionized biomolecular structure prediction, but developing and training new models remains a considerable challenge. To facilitate the development of new models, we present AtomWorks: a broadly applicable data framework for developing state-of-the-art biomolecular foundation models spanning diverse tasks, including structure prediction, generative protein design, and fixed backbone sequence design. We use AtomWorks to train RosettaFold-3 (RF3), a structure prediction network capable of predicting arbitrary biomolecular complexes with an improved treatment of chirality that narrows the performance gap between closed-source AlphaFold3 (AF3) and existing open-source implementations. We expect that AtomWorks will accelerate the next generation of open-source biomolecular machine learning models and that RF3 will be broadly useful as a structure prediction tool. To this end, we release the AtomWorks framework (https://github.com/RosettaCommons/atomworks), together with curated training data, code and model weights for RF3 (https://github.com/RosettaCommons/modelforge) under a permissive BSD license.
Figures



References
-
- Jumper John, Evans Richard, Pritzel Alexander, Green Tim, Figurnov Michael, Tunyasuvunakool Kathryn, Ronneberger Olaf, Bates Russ, Žídek Augustin, Bridgland Alex, et al. Alphafold 2. Fourteenth Critical Assessment of Techniques for Protein Structure Prediction, 2020.
-
- Baek Minkyung, DiMaio Frank, Anishchenko Ivan, Dauparas Justas, Ovchinnikov Sergey, Lee Gyu Rie, Wang Jue, Cong Qian, Kinch Lisa N, Schaeffer R. Dustin, Millán Claudia, Park Hahnbeom, Adams Carlos, Glassman Craig R, DeGiovanni Alexander, Pereira Jose H, Rodrigues Andrew V, van Dijk Amelie A, Ebrecht Andrea C, Opperman D. J., Sagmeister Tobias, Buhlheller Christoph, Pavkov-Keller Tea, Rathinaswamy Manoj K, Dalwadi Ujwala, Yip Christopher K, Burke John E, Garcia K. Christopher, Grishin Nick V, Adams Paul D, Read Randy J, and Baker David. Accurate prediction of protein structures and interactions using a three-track neural network. Science, 373(6557):871–876, 2021. doi: 10.1126/science.abj8754. URL https://www.science.org/doi/10.1126/science.abj8754. - DOI - PMC - PubMed
-
- Evans Richard, O’Neill Michael, Pritzel Alexander, Antropova Natasha, Senior Andrew, Green Timothy, Žídek Augustin, Bates Russell, Blackwell Sam, Yim Jason, Ronneberger Olaf, Bodenstein Sebastian, Zielinski Mirko, Bridgland Andrej, Potapenko Alexander, Cowie Charlie, Tunyasuvunakool Kathryn, Jain Ruchi, Clancy Ellen, Kohli Pushmeet, Jumper John, and Hassabis Demis. Protein complex prediction with alphafold-multimer. bioRxiv, page 2021.10.04.463034, 2022. doi: 10.1101/2021.10.04.463034. URL https://doi.org/10.1101/2021.10.04.463034. - DOI
-
- Krishna Rohith, Wang Jue, Ahern Woody, Sturmfels Pascal, Venkatesh Preetham, Kalvet Indrek, Lee Gyu Rie, Morey-Burrows Felix S, Anishchenko Ivan, Humphreys Ian R, et al. Generalized biomolecular modeling and design with rosettafold all-atom. Science, 384(6693): eadl2528, 2024. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
