This is a preprint.
The Data Distillery: A Graph Framework for Semantic Integration and Querying of Biomedical Data
- PMID: 40832351
- PMCID: PMC12363844
- DOI: 10.1101/2025.08.11.666099
The Data Distillery: A Graph Framework for Semantic Integration and Querying of Biomedical Data
Abstract
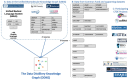
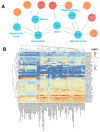
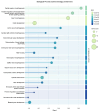
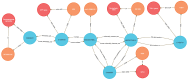
The Data Distillery Knowledge Graph (DDKG) is a framework for semantic integration and querying of biomedical data across domains. Built for the NIH Common Fund Data Ecosystem, it supports translational research by linking clinical and experimental datasets in a unified graph model. Clinical standards such as ICD-10, SNOMED, and DrugBank are integrated through UMLS, while genomics and basic science data are structured using ontologies and standards such as HPO, GENCODE, Ensembl, STRING, and ClinVar. The DDKG uses a property graph architecture based on the UBKG infrastructure and supports ontology-based ingestion, identifier normalization, and graph-native querying. The system is modular and can be extended with new datasets or schema modules. We demonstrate its utility for informatics queries across eight use cases, including regulatory variant analysis, tissue-specific expression, biomarker discovery, and cross-species variant prioritization. The DDKG is accessible via a public interface, a programmatic API, and downloadable builds for local use.
Figures



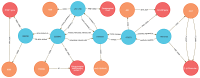
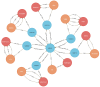
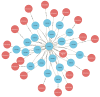
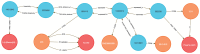
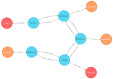
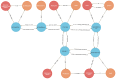
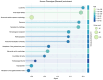
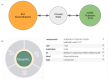
References
-
- Barabási A.-L. & Oltvai Z. N. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5, 101–113 (2004). - PubMed
-
- Alshahrani M., Thafar M. A. & Essack M. Application and evaluation of knowledge graph embeddings in biomedical data. PeerJ Comput Sci 7, e341 (2021).
-
- Alshahrani M. & Hoehndorf R. Drug repurposing through joint learning on knowledge graphs and literature. bioRxiv 385617 (2018) doi: 10.1101/385617. - DOI
Publication types
Grants and funding
- U01 AG072439/AG/NIA NIH HHS/United States
- OT2 OD030547/OD/NIH HHS/United States
- OT2 OD032092/OD/NIH HHS/United States
- U24 OD038422/OD/NIH HHS/United States
- OT2 OD030541/OD/NIH HHS/United States
- OT2 OD030546/OD/NIH HHS/United States
- U24 OD038423/OD/NIH HHS/United States
- OT2 OD030544/OD/NIH HHS/United States
- R03 OD030600/OD/NIH HHS/United States
- OT2 OD030545/OD/NIH HHS/United States
- OT2 OD030162/OD/NIH HHS/United States
- U24 CA264250/CA/NCI NIH HHS/United States
- U2C DK119886/DK/NIDDK NIH HHS/United States
- U01 CA200059/CA/NCI NIH HHS/United States
- OT2 OD032619/OD/NIH HHS/United States
- OT2 OD036435/OD/NIH HHS/United States
- OT2 OD033759/OD/NIH HHS/United States
- R24 GM146616/GM/NIGMS NIH HHS/United States
- U24 DK141185/DK/NIDDK NIH HHS/United States
- U24 CA268108/CA/NCI NIH HHS/United States
- U54 DA049098/DA/NIDA NIH HHS/United States
- U24 CA271114/CA/NCI NIH HHS/United States
- OT2 OD030160/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
