This is a preprint.
Large Language Models for Psychiatric Phenotype Extraction from Electronic Health Records
- PMID: 40832382
- PMCID: PMC12363723
- DOI: 10.1101/2025.08.07.25333172
Large Language Models for Psychiatric Phenotype Extraction from Electronic Health Records
Abstract
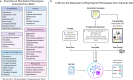
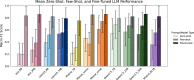
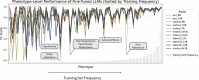
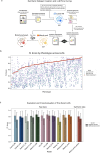
The accurate detection of clinical phenotypes from electronic health records (EHRs) is pivotal for advancing large-scale genetic and longitudinal studies in psychiatry. Free-text clinical notes are an essential source of symptom-level information, particularly in psychiatry. However, the automated extraction of symptoms from clinical text remains challenging. Here, we tested 11 open-source generative large language models (LLMs) for their ability to detect 109 psychiatric phenotypes from clinical text, using annotated EHR notes from a psychiatric clinic in Colombia. The LLMs were evaluated both "out-of-the-box" and after fine-tuning, and compared against a traditional natural language processing (tNLP) method developed from the same data. We show that while base LLM performance was poor to moderate (0.2-0.6 macro-F1 for zero-shot; 0.2-0.74 macro-F1 for few shot), it improved significantly after fine-tuning (0.75-0.86 macro-F1), with several fine-tuned LLMs outperforming the tNLP method. In total, 100 phenotypes could be reliably detected (F1>0.8) using either a fine-tuned LLM or tNLP. To generate a fine-tuned LLM that can be shared with the scientific and medical community, we created a fully synthetic dataset free of patient information but based on original annotations. We fine-tuned a top-performing LLM on this data, creating "Mistral-small-psych", an LLM that can detect psychiatric phenotypes from Spanish text with performance comparable to that of LLMs trained on real EHR data (macro-F1=0.79). Finally, the fine-tuned LLMs underwent an external validation using data from a large psychiatric hospital in Colombia, the Hospital Mental de Antioquia, highlighting that most LLMs generalized well (0.02-0.16 point loss in macro-F1). Our study underscores the value of domain-specific adaptation of LLMs and introduces a new model for accurate psychiatric phenotyping in Spanish text, paving the way for global precision psychiatry.
Conflict of interest statement
Competing Interests: All authors declare no financial or non-financial competing interests.
Figures
References
-
- Electronic health records and stratified psychiatry: bridge to precision treatment? | Neuropsychopharmacology. https://www.nature.com/articles/s41386-023-01724-y. - PMC - PubMed
-
- Optimising the use of electronic medical records for large scale research in psychiatry | Translational Psychiatry. https://www.nature.com/articles/s41398-024-02911-1. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
