Development of an All-Hydrocarbon Stapled Peptide Targeting BRK1 in Triple-Negative Breast Cancer
- PMID: 40832548
- PMCID: PMC12359003
- DOI: 10.1021/acsmedchemlett.5c00221
Development of an All-Hydrocarbon Stapled Peptide Targeting BRK1 in Triple-Negative Breast Cancer
Abstract
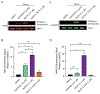
Wiskott-Aldrich syndrome protein family (WASF) members are key regulators of actin cytoskeleton dynamics at the leading edge of the cell membrane. WASF3 has been demonstrated to directly promote cancer invasion and metastasis in triple-negative breast cancer. WASF3 is incorporated into a heteropentameric protein complex with BRK1, CYFIP1/2, NCKAP1/1L, and ABI1/2/3 termed the WASF Regulatory Complex (WRC) that links upstream signaling pathways to Arp2/3-mediated actin nucleation. Disruption of the complex inhibits actin remodeling and presents a novel approach to targeting cancer invasion and metastasis. Here we report the development of a first-generation all-hydrocarbon stapled BRK1 mimetic peptide, BASH-2, designed to inhibit BRK1 binding within the WRC to disrupt proper WRC assembly and function. BASH-2 was found to permeate cells, bind to WASF3 and ABI2, and inhibit cancer cell migration and invasion in a dose-dependent manner. BASH-2 may present a novel approach to targeting WASF3-promoted invasion and metastasis in triple-negative breast cancer.
Keywords: BRK1; Stapled peptide; WASF Regulatory Complex; WASF3; triple-negative breast cancer.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Kulkarni S; Augoff K; Rivera L; McCue B; Khoury T; Groman A; Zhang L; Tian L; Sossey-Alaoui K, Increased expression levels of WAVE3 are associated with the progression and metastasis of triple negative breast cancer. PLoS One 2012, 7 (8), e42895. DOI: 10.1371/journal.pone.0042895. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

