The role of cyclic di-GMP in biomaterial-associated infections caused by commensal Escherichia coli
- PMID: 40833985
- PMCID: PMC12367115
- DOI: 10.1371/journal.pone.0330229
The role of cyclic di-GMP in biomaterial-associated infections caused by commensal Escherichia coli
Abstract
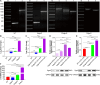
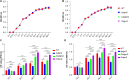
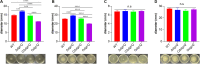
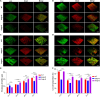
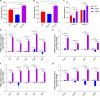
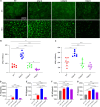
Biofilms are protective structures that bacteria use to evade the immune system and resist antibiotics, leading to complications in medical treatments, especially with implanted devices. The molecule cyclic di-GMP (c-di-GMP) is crucial for biofilm formation in Escherichia coli (E. coli). To understand its role in biomaterial-associated infections (BAIs), we created four E. coli strains with varying c-di-GMP levels: a knockout strain (ΔdgcQ), an overexpression strain (OdgcQ), a complemented strain (CΔdgcQ), and a wild-type mutant strain (WT). By employing in vitro BAI models and techniques such as crystal violet (CV) staining, XTT assay, confocal laser scanning microscopy (CLSM), and scanning electron microscopy (SEM), we observed that the ΔdgcQ strain, with low c-di-GMP levels, adhered more readily to biomaterial surfaces at the initial stage of biofilm formation, yet faced difficulties in sustaining mature biofilms. In contrast, OdgcQ and CΔdgcQ with higher c-di-GMP were able to generate more mature biofilms on biomaterial surfaces. Additionally, c-di-GMP was found to negatively regulate bacterial swimming motility and enhance the ability to cope with environmental stresses. The results also reiterate the canonical function of c-di-GMP, which is to reduce the motility of bacteria. Concurrently, gene expression analysis confirmed these findings, revealing that genes related to motility (flhC, flhD, motA, motB, ycgR), extracellular polymeric substances (EPS) synthesis (csgA, csgD, bcsA, ynfM), and stress resistance (sodA, katE, rstA, ibpA, ibpB, hdeA, hdeD, gadA, gadB) were consistently up-regulated in OdgcQ with high c-di-GMP levels. Importantly, ΔdgcQ considerably promoted the adhesion to and invasion of host cells and elicited a stronger host immune response, whereas OdgcQ impaired the ability to interact with host cells, as evidenced by decreased adhesion/invasion and inhibited release of inflammatory cytokines (IL-1β, IFN-β, IP-10, and NF-κB). Collectively, our findings shed light on the c-di-GMP signaling pathway's role in BAIs and propose that modulating this pathway could be a promising strategy for combating E. coli-induced BAIs.
Copyright: © 2025 Li et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Mori T, Yamamoto H, Tabata T, Shimizu T, Endo Y, Hanasawa K, et al. A free radical scavenger, edaravone (MCI-186), diminishes intestinal neutrophil lipid peroxidation and bacterial translocation in a rat hemorrhagic shock model. Crit Care Med. 2005;33(5):1064–9. doi: 10.1097/01.ccm.0000162952.14590.ec - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

