Common molecular links and therapeutic insights between type 2 diabetes and kidney cancer
- PMID: 40834023
- PMCID: PMC12367126
- DOI: 10.1371/journal.pone.0330619
Common molecular links and therapeutic insights between type 2 diabetes and kidney cancer
Abstract
Introduction: Type 2 diabetes (T2D) is considered as a risk factor for kidney cancer (KC). However, so far, there is no study in the literature that has explored genetic factors through which T2D drive the development and progression of KC. Therefore, this study attempted to explore T2D- and KC-causing shared key genes (sKGs) for revealing shared pathogenesis and therapeutic drugs as their common treatments.
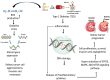
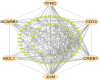
Methods: The integrated bioinformatics and system biology approaches were utilized in this study. The statistical LIMMA approach was used based web-tool GEO2R to detect differentially expressed genes (DEGs) through transcriptomics analysis. Then upregulated and downregulated DEGs for T2D and KC were combined to obtained shared DEGs (sDEGs) between T2D and KC. The STRING database was used to construct the protein-protein interaction (PPI) network of sDEGs. Then Cytohubba plugin-in Cytoscape were used in the PPI network to disclose the sKGs based on different topological measures. The RegNetwork database was used in NetworkAnalyst to analyze co-regulatory networks of sKGs with transcription factors (TFs) and micro-RNAs to identify key TFs and miRNAs as the transcriptional and post-transcriptional regulators of sKGs, respectively. AutoDock Vina is a tool used for molecular docking. ADME/T properties were 24 assessed using pkCSM and SwissADME.
Results: At first, 74 shared DEGs (sDEGs) were identified that can distinguish both KC and T2D patients from control samples. Through protein-protein interaction (PPI) network analysis, top-ranked 6 sDEGs (CD74, TFRC, CREB1, MCL1, SCARB1 and JUN) were detected as the sKGs that drive both KC and T2D development and progression. The most common sKG 'CD74' is associated with key pathways, such as NF-κB signaling transduction, apoptotic processes, B cell proliferation. Differential expression patterns of sKGs validated by independent datasets of NCBI database for T2D and TCGA and GTEx databases for KC. Furthermore, sKGs were found to be significant at several CpG sites in DNA methylation studies. Regulatory network analysis identified three TFs proteins (SMAD5, ATF1 and NR2F1) and two miRNAs (hsa-mir-1-3p and hsa-mir-34a-5p) as the regulators of sKGs. The enrichment analysis of sKGs with KEGG-pathways and Gene Ontology (GO) terms revealed some crucial shared pathogenetic mechanisms (sPM) between two diseases. Finally, sKGs-guided four potential therapeutic drug molecules (Imatinib, Pazopanib hydrochloride, Sorafenib and Glibenclamide) were recommended as the common therapies for KC with T2D.
Conclusion: The results of this study may be useful resources for the diagnosis and therapy of KC with the co-existence of T2D.
Copyright: © 2025 Ahmmed et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Bioinformatics analysis to disclose shared molecular mechanisms between type-2 diabetes and clear-cell renal-cell carcinoma, and therapeutic indications.Sci Rep. 2024 Aug 19;14(1):19133. doi: 10.1038/s41598-024-69302-w. Sci Rep. 2024. PMID: 39160196 Free PMC article.
-
Unveiling the Potential Role of Hesperetin and Emodin as a Combination Therapy to Inhibit the Pancreatic Cancer Progression against the C-Met Gene.Protein Pept Lett. 2025;32(4):280-298. doi: 10.2174/0109298665363165250225100109. Protein Pept Lett. 2025. PMID: 40129158
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

