Daxx mediated histone H3.3 deposition on HSV-1 DNA restricts genome decompaction and the progression of immediate-early transcription
- PMID: 40834051
- PMCID: PMC12393724
- DOI: 10.1371/journal.ppat.1012501
Daxx mediated histone H3.3 deposition on HSV-1 DNA restricts genome decompaction and the progression of immediate-early transcription
Abstract
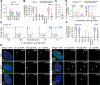
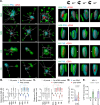
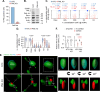
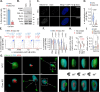
Herpesviruses are ubiquitous pathogens that cause a wide range of disease. Upon nuclear entry, their genomes associate with histones and chromatin modifying enzymes that regulate the progression of viral transcription and outcome of infection. While the composition and modification of viral chromatin has been extensively studied on bulk populations of infected cells by chromatin immunoprecipitation, this key regulatory process remains poorly defined at single-genome resolution. Here we use high-resolution quantitative imaging to investigate the spatial proximity of canonical and variant histones at individual Herpes Simplex Virus 1 (HSV-1) genomes within the first 90 minutes of infection. We identify significant population heterogeneity in the stable enrichment and spatial proximity of canonical histones (H2A, H2B, H3.1) at viral DNA (vDNA) relative to established promyelocytic leukaemia nuclear body (PML-NB) host factors that are actively recruited to viral genomes upon nuclear entry. We show the replication-independent histone H3.3/H4 chaperone Daxx to cooperate with PML to mediate the enrichment and spatial localization of variant histone H3.3 at vDNA and limit the rate of HSV-1 genome decompaction. This host response is counteracted by the viral ubiquitin ligase ICP0, which degrades PML to disperse Daxx and variant histone H3.3 from vDNA to stimulate the progression of viral immediate-early (IE) transcription, genome expansion, and onset of HSV-1 replication. Our data support a model of intermediate and sequential histone assembly initiated by Daxx that limits the rate of HSV-1 genome decompaction independently of the stable enrichment of histones H2A and H2B at vDNA required to facilitate canonical nucleosome assembly. We identify HSV-1 genome decompaction upon nuclear infection to play a key role in the initiation and functional outcome of HSV-1 lytic infection, findings pertinent to the transcriptional regulation of many nuclear replicating herpesvirus pathogens.
Copyright: © 2025 Roberts et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Update of
-
Daxx mediated histone H3.3 deposition on HSV-1 DNA restricts genome decompaction and the progression of immediate-early transcription.bioRxiv [Preprint]. 2024 Aug 15:2024.08.15.608064. doi: 10.1101/2024.08.15.608064. bioRxiv. 2024. Update in: PLoS Pathog. 2025 Aug 20;21(8):e1012501. doi: 10.1371/journal.ppat.1012501. PMID: 39185184 Free PMC article. Updated. Preprint.
Similar articles
-
Daxx mediated histone H3.3 deposition on HSV-1 DNA restricts genome decompaction and the progression of immediate-early transcription.bioRxiv [Preprint]. 2024 Aug 15:2024.08.15.608064. doi: 10.1101/2024.08.15.608064. bioRxiv. 2024. Update in: PLoS Pathog. 2025 Aug 20;21(8):e1012501. doi: 10.1371/journal.ppat.1012501. PMID: 39185184 Free PMC article. Updated. Preprint.
-
Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication.J Virol. 2018 Apr 27;92(10):e00057-18. doi: 10.1128/JVI.00057-18. Print 2018 May 15. J Virol. 2018. PMID: 29491153 Free PMC article.
-
Promyelocytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/Histone H3.3/H3.3 Chaperone Axis.PLoS Pathog. 2018 Sep 20;14(9):e1007313. doi: 10.1371/journal.ppat.1007313. eCollection 2018 Sep. PLoS Pathog. 2018. PMID: 30235352 Free PMC article.
-
Daxx and HIRA go viral - How chromatin remodeling complexes affect DNA virus infection.Tumour Virus Res. 2025 Jun;19:200317. doi: 10.1016/j.tvr.2025.200317. Epub 2025 Mar 20. Tumour Virus Res. 2025. PMID: 40120981 Review.
-
The Dynamic Regulation of Daxx-Mediated Transcriptional Inhibition by SUMO and PML NBs.Int J Mol Sci. 2025 Jul 12;26(14):6703. doi: 10.3390/ijms26146703. Int J Mol Sci. 2025. PMID: 40724953 Free PMC article. Review.
References
-
- Knipe DM, Howley PM. Fields virology. 6th ed. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health. 2013.
-
- Davison AJ. Comparative analysis of the genomes. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R. Human herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge. 2007. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

