Efficient termination of transcription by RNA polymerase I requires a conserved hairpin of the ribosomal RNA precursor
- PMID: 40834087
- PMCID: PMC12366685
- DOI: 10.1126/sciadv.adw2470
Efficient termination of transcription by RNA polymerase I requires a conserved hairpin of the ribosomal RNA precursor
Abstract
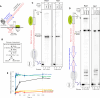
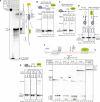
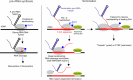
RNA polymerase I (Pol I) synthesizes ribosomal RNA precursor (pre-rRNA), which comprises most of RNA in eukaryotic cells. Despite decades of investigation, there is still no consensus on what causes Pol I transcription termination. Here, we show that efficient termination by Pol I, paused by termination roadblock protein, is caused by RNA hairpin of the nascent pre-rRNA. Hairpin-dependent termination takes place at a physiological rate and does not require trans-acting factors. The function of the roadblock protein and the T-rich sequence is to synergistically cause deep backtracking of Pol I toward the termination RNA hairpin. Simultaneously, Pol I is catalytically inactivated, preventing rescue from backtracking through RNA cleavage and thus committing Pol I to termination. Termination RNA hairpins are present in most of Pol I terminators of eukaryotes, suggesting conservation of the RNA hairpin-dependent mechanism of Pol I transcription termination. We propose a simple model that unifies previous findings.
Figures
Similar articles
-
High-resolution Sequencing Reveals that the Paf1 Complex May be a Conserved Transcription Elongation Factor for Eukaryotic RNA Polymerase I.J Mol Biol. 2025 Sep 1;437(17):169220. doi: 10.1016/j.jmb.2025.169220. Epub 2025 May 19. J Mol Biol. 2025. PMID: 40398673
-
Specific DNA features of the RNA polymerase I core promoter element targeted by core factor.Biochim Biophys Acta Gene Regul Mech. 2025 Jun;1868(2):195088. doi: 10.1016/j.bbagrm.2025.195088. Epub 2025 Apr 9. Biochim Biophys Acta Gene Regul Mech. 2025. PMID: 40216226
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
The ribosomal RNA transcription landscapes of Plasmodium falciparum and related apicomplexan parasites.Nucleic Acids Res. 2025 Jul 8;53(13):gkaf641. doi: 10.1093/nar/gkaf641. Nucleic Acids Res. 2025. PMID: 40626563 Free PMC article. Review.
-
Transcription factor regulation of ribosomal RNA in hematopoiesis.Curr Opin Hematol. 2024 Jul 1;31(4):199-206. doi: 10.1097/MOH.0000000000000816. Epub 2024 Apr 11. Curr Opin Hematol. 2024. PMID: 38568093 Free PMC article. Review.
References
-
- Merkl P., Perez-Fernandez J., Pilsl M., Reiter A., Williams L., Gerber J., Bohm M., Deutzmann R., Griesenbeck J., Milkereit P., Tschochner H., Binding of the termination factor Nsi1 to its cognate DNA site is sufficient to terminate RNA polymerase I transcription in vitro and to induce termination in vivo. Mol. Cell Biol. 34, 3817–3827 (2014). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

